Improvement
Improvement for BBa_K523013(Plac+INP-EYFP)
Design
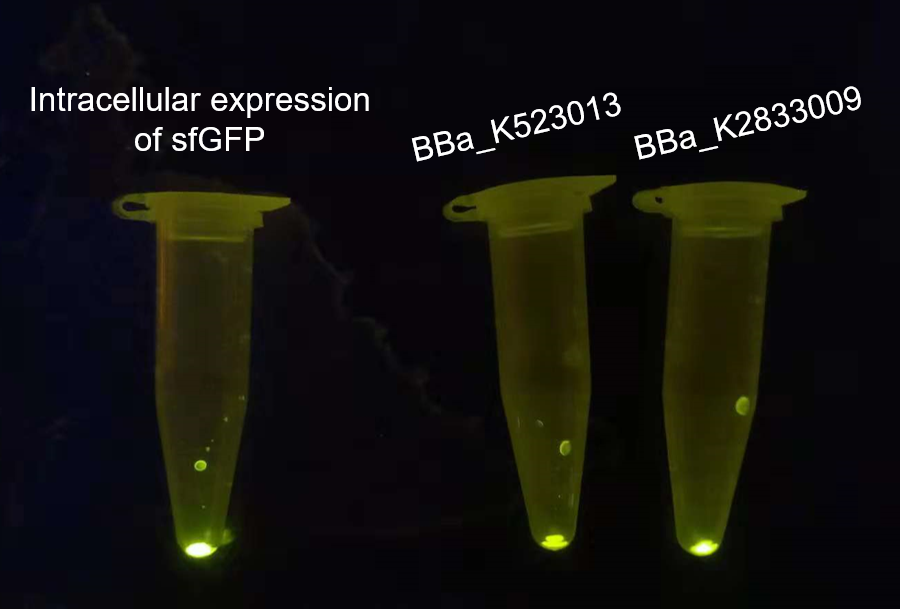
Our project used surface display system, and we chose the INP-YFP expressing part BBa_K523013 built by Edinburgh 2011 to construct the INP-mediated surface display system for our interest protein. While testing the surface display efficiency of this part, we found that the fluorescence intensity was too weak both under the uv-light(adjusted to the same OD600)(Fig.2, middle) and under the super resolution microscope(Fig.3, middle).
To characterize the surface display efficiency of INP better, we added a strong promoter BBa_J23104 and RBS BBa_B0032 to the upstream of INP, and changed the YFP to sfGFP, to construct our parts BBa_K2833009(Fig.1)

Results
The overnight cultured bacteria expressing GFP, INP-YFP and INP-GFP respectively were firstly adjusted to OD600 = 1.0 to assure the same cell counts, then 1 mL bacteria solution of each was centrifuged to get the sediment of whole cell. From the fluorescence strength of the whole cell(E.coli, DH5α) precipitation under UV-light we can see that the BBa_K523013 transformed cells showed the weakest fluorescence, suggesting that our new construct could give out stronger fluorescence.
We observed the fluorescence under the super-resolution microscope(N-SIM). As shown in figure 3, The intracellular expression of GFP displayed the rode-shape of E.coli(fig.3 left), while the signal of surface displayed GFP and YFP showed the dotted pattern(figure 3, middle, right), which suggested that the GFP and YFP were gathered and distributed on the surface of E.coli.. From this result we can see that fluorescence signal was weaker in the BBa_K523013 transformed cells than in BBa_K2833009 transformed cells, suggesting that our new construct could make the INP-mediated surface display system more visible.

Improvement for BBa_K1321200(J23101+B0034+VHb)
Design
Our project aimed to use Vitreoscilla hemoglobin (VHb) to improve the oxygen usage of the engineering bacteria. iGEM14_Imperial has constructed BBa_K1321200 which can express VHb under the control of J23101 and B0032. However, when making alignment we found that the sequence of VHb in BBa_K1321200 was different from the original VHb both in DNA and protein. (data source: NCBI)

So we wondered which VHb could affect the growth of bacteria on a higher level. We constructed BBa_K2833022 and BBa_K2833023 with different promoters as shown in figure 5.

Results
We transformed these constructs to E.coli.(BL21) and measured their growth curve(figure 6). From the result we can see that all the three VHb transgene bacteria grew better than the wildtype bacteria, mainly presented on the maximal bio-mass at platform, while our newly built constructs(BBa_K2833022, BBa_K2833023) showed bigger effects on the growth of the bacteria.
However, the difference of the growth curves was not very significant, and we thought maybe the normal condition contained enough oxygen so that the bacteria can grow well themselves. So in the future, we are supposed to optimize the experimental condition such as creating a low-oxygen content environment and so on.

