Collaboration
FJNU-2018
1.Help FJNU-2018 to test the antibacterial ability of PLA
We helped FJNU-2018 to inhibit the strains of Staphylococcus aureus and Bacillus subtilis by PLA, and verified the broad-spectrum antibacterial ability of PLA.Our two teams often communicate with each other over the summer, during which we exchanged experience and interesting things through chat tools in daily life and establish a good relationship.
2.Our two teams often communicate with each other over the summer, during which we exchanged experience and interesting things through chat tools in daily life and establish a good relationship.
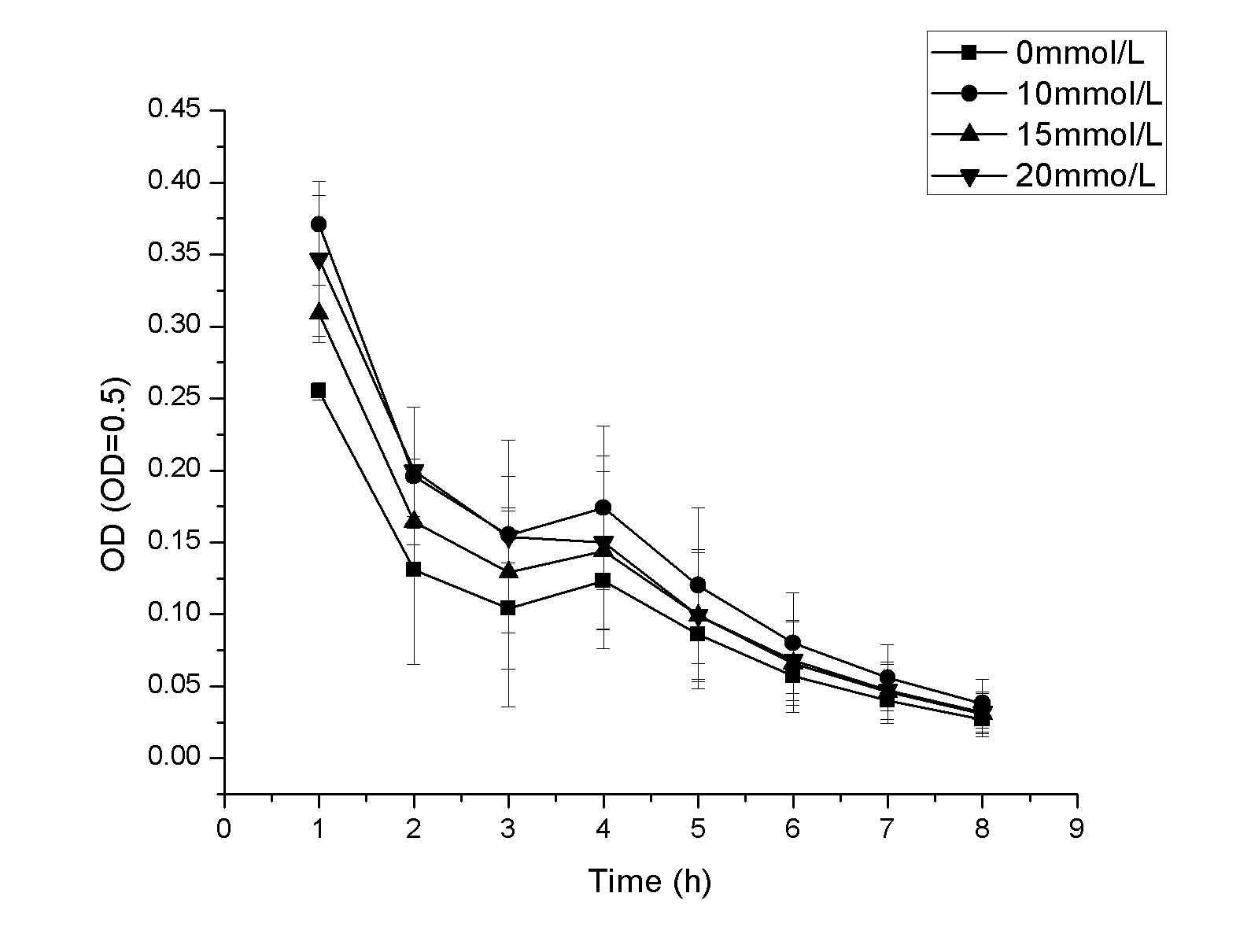
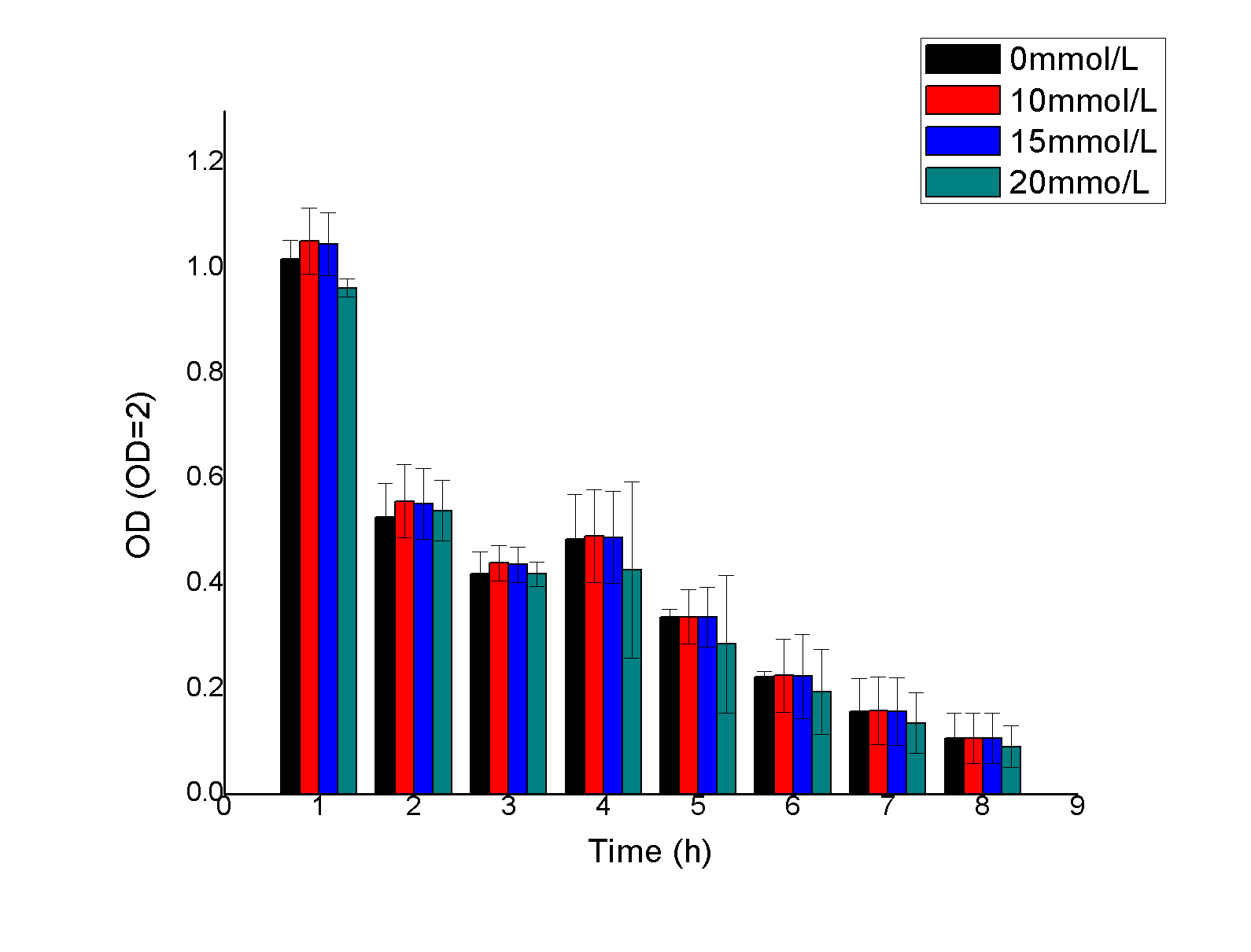
3、FJNU-2018 helped us to measuresome data of ingredients in the soil.
they used a five point sampling method to sample the soil in our team's area and shared data with us. Details can be found on this page:https://2018.igem.org/Team:FJNU-China/Collaborations.

ECUST
1. Help ECUST to carry out their mathematical modeling
We helped ECUST to build their quorum sensing model and anti-biofilm activity model, and provided corresponding guidance for it, and proposed a new mathematical modeling idea.
2.ECUST helped us to measuresome data of ingredients in the soil
They used a five point sampling method to sample the soil in our team's area and shared data with us.
Details can be found on this page:https://2018.igem.org/Team:ECUST/Collaborations
OUC-China
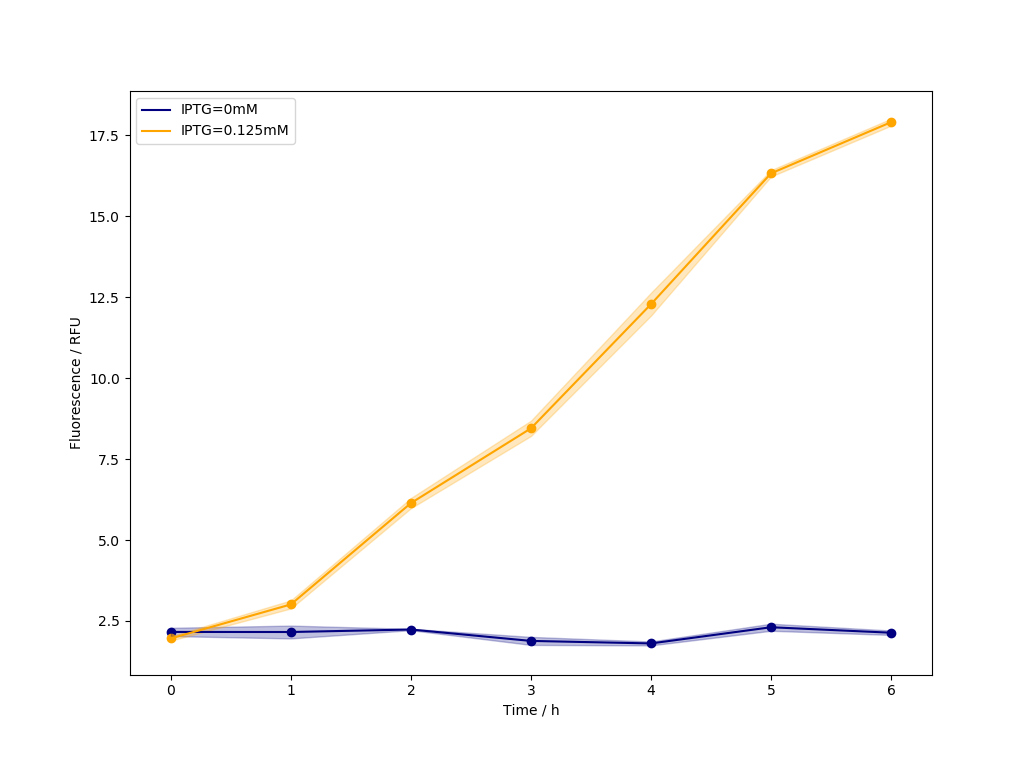
1.Help OUC-China to test miniToe system
We are so interested in miniToe system,and with the material provided by them,we successfully proved their system.
2.OUC-China provided us with samples of salt-containing wastewater from local factories, which helped us to modify the direction of the project and test the ability of the target strain to work under complex water quality conditions.
CPU-China
1.Help CPU-CHINA to model
We provided C++ programming code to CPU-CHINA to help its model better predict the effect of treatment on patients.
2. CPU-CHINA provided us with samples of salt-containing wastewater from local factories, which helped us to modify the direction of the project and test the ability of the target strain to work under complex water quality conditions.
NPU-China
Using their laboratory's unique Automation Enzyme Mutation Design (AEMD) system, NPU-China helped us analyze the core enzyme of the project, the KcsA protein, and designed the enzyme modification from both functional and stability aspects. When we modified KcsA, we partially referred to this data.
CCIC
We participated in the CCIC sponsored by ShanghaiTech in 2018. At the conference we gained a lot of knowledge about synthetic biology and many novel ideas, and learned a lot about other team projects. In the exchange, we met new friends and laid the foundation for future cooperation.

Meet with the UESTC-China and SCU-China
We met at the Sichuan University School of Life Sciences with the UESTC-China and SCU-China. We exchanged their projects and conducted some technical discussions and research. We also got guidance on protein expression.

Meeting withNPU-China and XJTU-China
We met at NPU-China and XJTU-China at the College of Life Sciences of Northwestern Polytechnical University. At the meeting, we exchanged the contents of the project and exchanged ideas on team building and experience.

Finally, thanks to CPU-China and OUC-China for collecting water samples for us. And thanks to FJNU-China and ECUST for providing us with local soil salt content data. Their work provides a good reference for us to study the practical application value, application scope and market demand of this project.

