Microfluidic system
1) Double emulsion system
We developed two systems for the same purpose to have two single cells cocultured in thousands of separated microenvironments so as to have thousands of reactions at the same time which is time-saving. One system is double emulsion system and the other is micro-well system.

Figure 1 Scheme of double emulsion system. (Green and red dots represent cells)
As shown in Figure 1, there are two steps in double emulsion system. In the first step, two type of cells from two channels flow into the chip. Oil will cut the aqueous cell medium generating many water-in-oil droplets with size is around 30 microns. The encapsulation principle is mainly due to hydrophobic effect of oil and water. The second emulsion will form the water-in-oil-water droplets which can be used in Fluorescence-activated Cell Sorting (FACS).
Explore more about double emulsion system.
2) Micro-well system

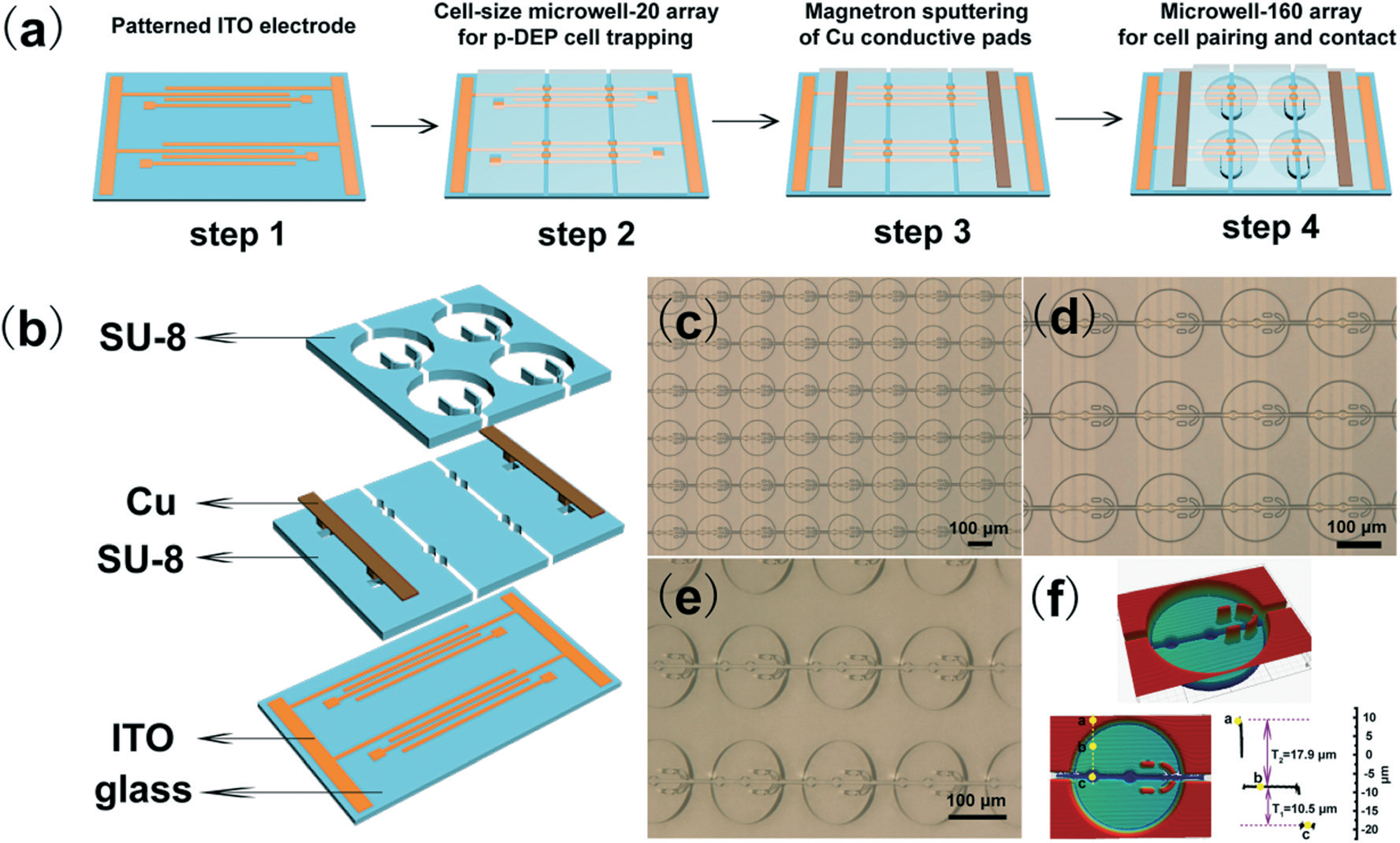
Figure 2 Scheme of micro-well planar system
As shown in Figure 2, micro-well system can capture two types of cell in order. There are two 10-15micron wells in a 100micron well. Two cells can be captured by electrostatic force in to two wells and react separately in the well. They will be isolated into thousands of microenvironment for signal transduction. After reaction, we can pick up the cells with capillary tubes under fluorescence microscope.
Explore more about micro-well system.
Biology
1) Wnt secretion and signaling pathway

Figure 3 Wnt secretion and Wnt signaling pathway
Canonical Wnt pathway plays important role in embryonic development and cancer initiation. In Wnt secreting cells, Wnt3A protein is transcribed into mRNA, which is translated into premature Wnt3A. Wnt3A protein is add a palmate tail at the C-terminal during post translational modification by a series of enzymes in ER and Golgi. It is then transported via vesicles to be secreted into the extracellular environment.
After secretion, Wnt responding cell will receipt the Wnt3A signal and cause downstream effect. The LRP5/6 and Frizzled will get close together due to presence of Wnt3A, which will induce phosphorylation of LRP5/6. And then, disruption complex will be fixed in the membrane and β-catenin protein is released. The concentration of β-catenin is increased and it enters nucleus to bind to TCF promoter and activates the downstream genes expression(GFP as a downstream target in Figure 3).
We mainly build a Wnt-secreting cell line using Mouse Lwnt3A cells and a Wnt-responding cell line using human HEK 293R cells.
2) CRISPR-Cas9 gene knockout

Figure 4 Principle of CRISPR-Cas9 knockout
After Cas9 protein and gRNA sequence are integrated into genome of host cells though Lentiviral infection, expressed Cas9 protein can cut the target DNA sequence complementary to gRNA sequence, which can introduce loss-of-function mutations in the gene after DNA random repair.
Due to the rapid development of CRISPR-Cas9 system, one gRNA can be inserted to one host cell in order to cut one specific gene guided by this gRNA. So it is able to have multiple genes knockout and even genome-wide knockout in multiple cells in one cell population with one specific gene knockout in a single cell.
Applying this method, we can parallelly generate thousands of single perturbations in genetic level. Genetic screening is our final goal for our system. In the experiment, we will knockout several genes (protocols) whose functions are known as proof-of-principle for our novel system.
3) Lentivirus transfection

Figure 5 Three lentiviral vectors for lentivirus production
Lentiviral vectors for cell transfection is widely used in mammalian cell lines. The second generation lentiviral vectors consists of three separate plasmids: packaging plasmid (psPAX2), envelop plasmid (pMD2.G) and a transfer plasmid.

Figure 6 Virus transfection
Lentivirus is produced by HEK 293T cell line and is then transfected into our target cell lines. The genetic construct in Lentiviral vector can be stably integrated into the genome of mammalian cells. And after antibiotic selection or other selection methods, a new cell line can be generated. The procedure of cell transfection contain plasmid design, virus production, cell transfection and positive selection. Safety is concerning during virus production.
In our experiments, lentiviral vectors were transformed into mouse and human cell lines to generate target cell line. Plasmid design and cell line construction please refer to Results-Engineered cells.
Observable equipment
1) Fluorescence microscope
Fluorescence microscope is for observation of microdroplets and micro-well plate. We can observe the droplet under fluorescence microscope and calculate the encapsulation rate for both microfluidic system.

Figure 7 Fluorescence microscope
2) Fluorescence-activated Cell Sorter(FACS)

Figure 8 Workflow of FACS
After encapsulation of two cells, cells will be coculture for a period of time. We can select microdroplets based on the fluorescence emitted from reporter cells.
The size of double emulsion droplets is about 30micron, which can be run in Fluorescence-activated cell sorter(FACS). Droplets are electrically charged based the results of analysis fluorescence. Sorting occurs then via deflection of the drop with the cell in an electrical field in different collection tubes. So, the strength of Wnt signal can be visualized and the droplet can be selected according to fluorescence threshold.
For micro-well plate, two ways to select the microwell. One is to pick up by capillary tubes under fluorescence microscope and the other is to fuse the secreting cell and reporter cell together to make a fusion cell. The fusion cell might have the fluorescence of downstream Wnt signal and bring the gene knockout information from secreting cell. And then, FACS can be used to select the fusion cell with different fluorescence intensity.
Next generation DNA sequencing

Figure 9 Flowchart of next generation sequencing
After selection, we need to extract the gene which is knockout before encapsulation. The next generation DNA sequencing will be applied to find the key genes in Wnt secretion pathway. A pair of primer on both sides of gRNA sequence are designed and enrich the gRNA sequence using Polymerase Chain Reaction(PCR). Our selected droplet have multiple gRNA so we need Illumina Sequencing to find out all the enriched gene sequence.
Finally, key genes in Wnt secretion will be screened out and listed according to its copy number in sequencing data. We can confirm their functions via one-by-one experiments.
At the same time, we use data from TCGA to predict the key gene in Wnt signaling as our comparative method in Modeling.


