Cloning
In our current project, we planned to clone two genes of interest, namely spaP and gtfC from Streptococcus mutans, into the standard psB1C3 plasmid. In addition, we designed our own promoter to induce the expression of our proteins. We utilized E. coli NEB Turbo to clone the genes and then transformed BL21 E. coli to express those genes.
Contents
Purpose of cloning
The aim of our project is to obtain aptamers that specifically bind to pivotal proteins of the biofilm formation by S. mutans. To this end, we cloned gtfC and spaP under the control of our trc promoter into psB1C3. Moreover, we cloned RFP under the control of the trc promoter into psB1C3.
Genes of interest
As described before, we use antigen I/II and the GTFC (also known as glycosyltransferase-SI) as targets to find binding aptamers against. The genes encoding them are called spaP and gtfC, respectively.
spaP
The gene encoding antigen I/II is called spaP.
Antigen I/II binds to the pellicle on the surface of our teeth and serves as an initial anchor for the binding of S. mutans. One specific receptor, the innate immunity scavenger receptor glycoprotein-340 (gp-340), has been found to be the main receptor for spaP. Interestingly, different glycosylations of gp-340 determine if it is present in a fluid or a surface-bound form. The fluid form leads to the aggregation of S. mutans and the clearance through swallowing, whereas the surface-bound form promotes the initial attachment of S. mutans to the tooth surface. Differential glycosylation patterns of gp-340 in human cohorts may contribute to different susceptibilities to caries.¹’²
Antigen I/II has been used in many vaccination studies against caries, which is why we target it with aptamers.³’⁴
The amino acid sequence for the protein can be found on Uniprot under the ID: P23504 and the gene for spaP can be found under the GeneID: 1028055 in NCBI.
gtfC
The family of glycosyltransferases of S. mutans is subdivided into 3 different GTFs, namely glycosyltransferase-I (GTFB), glycosyltransferase-SI (GTFC) and glycosyltransferase-S (GTFD). They all vary in their binding-capacity to either saliva-coated hydroxyapatite (sHA) or bacterial surfaces and the branching and solubility of the produced glucans.
GTFB mainly adheres to the surface of different bacteria whereas it does not bind to sHA. GTFC has a high affinity for sHA, but does not bind bacterial surfaces. GTFD binds to both surfaces with only very low affinities.⁵’⁶
The glucans produced by these glycosyltransferases can be subdivided into soluble and insoluble. GTFB mainly produces insoluble glucans with α-1-3-linkages, whereas GTFD only produces soluble glucans. GTFC is able to synthesize both soluble and insoluble glucans. Interestingly, the presence of starch hydrolytes may influence the branching of glucans produced by GTFB, whereas the other two glycosyltransferases are not affected by the products of starch hydrolysis by β-amylase.⁶’⁷
We focused on GTFC as this GTF binds to the pellicle of the teeth and targeting it may thus lead to a more long-lasting effect of the aptamers as a preventive mean against caries.
The amino acid sequence for GTFC can be found in Uniprot under the ID: P13470 and the gene for gtfC can be found under the GeneID: 1028343 in NCBI.
Approach
First, we codon-optimized the genes spaP and gtfC from S. mutans encoding antigen I/II and GTFC, so they can be easily transcribed and translated by E. coli. For this, we used the IDT codon optimization tool (https://eu.idtdna.com/CodonOpt).
Subsequently, we edited the sequences with alternative codons to eliminate the restriction sites for EcoRI, XbaI, PstI and SpeI. Moreover, we added a HindIII restriction site in the middle of the gtfC gene to be able to let it synthesize in two parts and then ligate the two parts together by the overlaps of the digested HindIII site. For spaP, we added a NheI restriction site in the middle to let it synthesize in two part and ligate them together.
Finally, we added the prefix and suffix upstream and downstream of every part. For the upstream parts, we always also set the trc promoter construct (BBa_K2402000) upstream of the ATG codon of the protein.
The cloning strategy for gtfC is indicated in Figure 1. The cloning strategy for spaP is indicated in Figure 2.
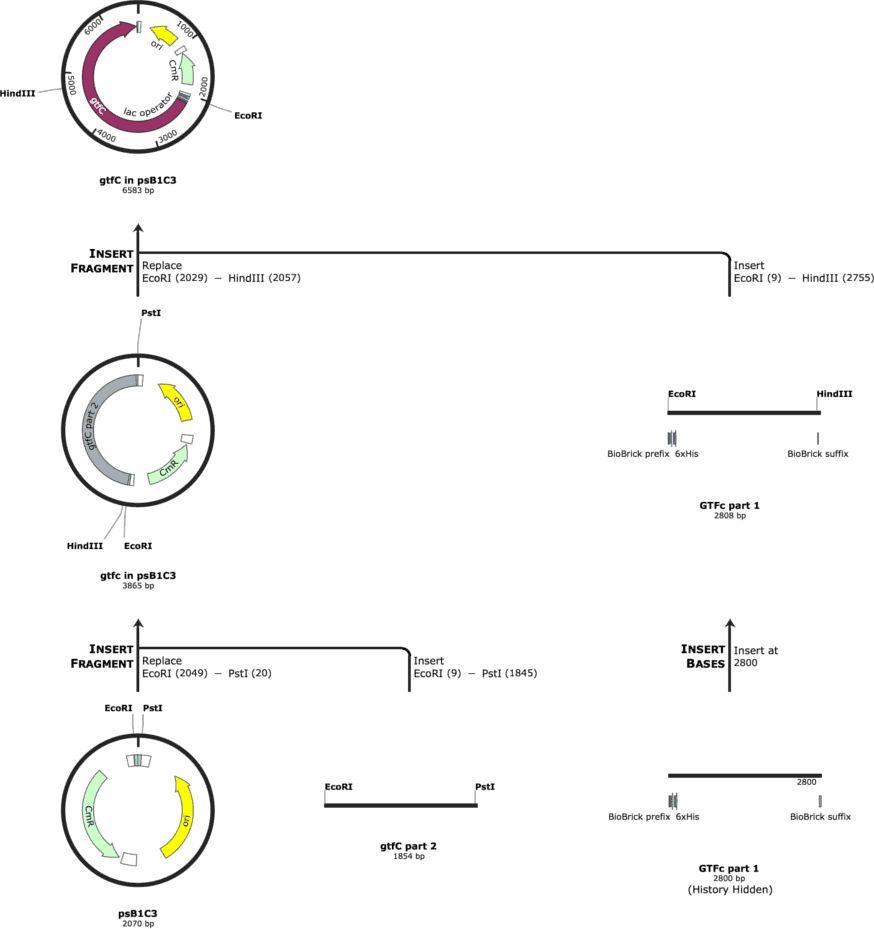
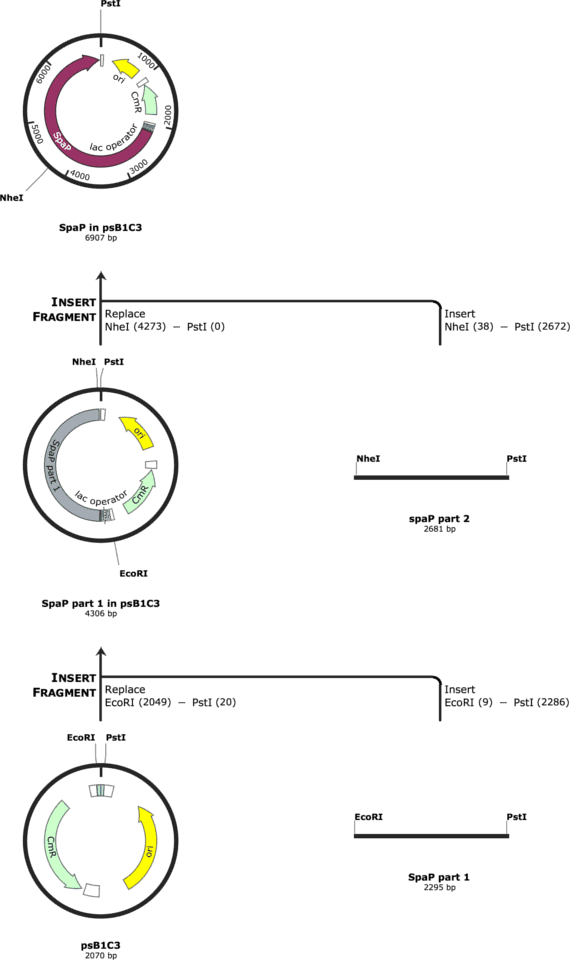
We had the parts synthesized by IDT and tried to ligate them into the psB1C3. The cloning was performed in NEB Turbo Competent E. coli and the protein expression was to be performed in BL21 E. coli.
Cloning
We were able to ligate gtfC part 2 (figure 3) and spaP part 1 (figure 4) into psB1C3.
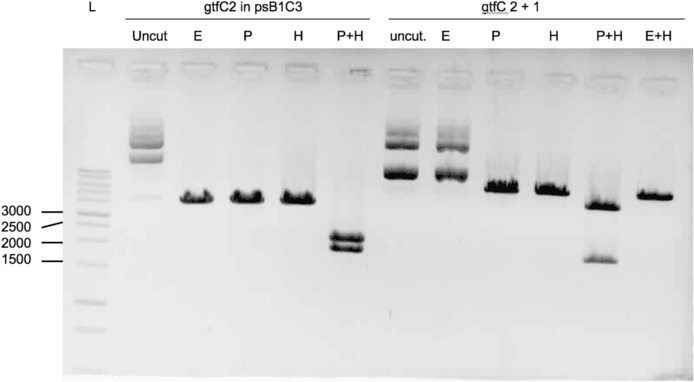
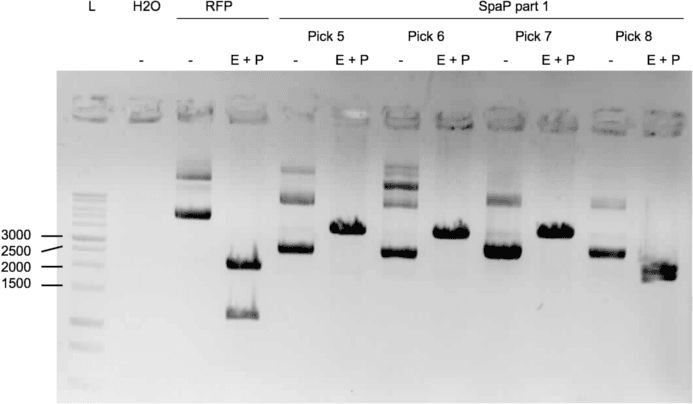
Protocols used
Restriction
When working with the DNA stock from IDT:
Restriction of gene of interest:
2,5 U EcoRI, 2,5 U PstI, 0,1 µl BSA, 1 µl Buffer H, 60ng DNA, add 10 µl ddH₂0
-> Incubated for 1h at 37°C, heat inactivation: 15 min at 65°C
Restriction of linearlized psB1C3:
2,5 U EcoRI, 2,5 U PstI, 2,5 U DpnI, 0,1 µl BSA, 1 µl Buffer B, 60ng DNA, add 10 µl ddH₂0
-> Incubated for 1h at 37°C, heat inactivation: 15 min at 65°C
When working with PCR amplified DNA or plasmids from minipreps, the amount of DNA was adjusted to 1 µg of DNA and 10 U of restriction enzyme.
For restriction analysis, 200ng of DNA were restricted with 2,5 U of the respective restriction enzymes in an end volume of 10 µl.
Dephosphorylation
Thermo Scientific Fast AP Thermosensitive Alkaline Phosphatase - Protocol for nucleic acid dephosphorylation
Ligation
For ligation we used the T4 DNA ligase by NEB according to the manufacturer's manual using 50ng of vector plus the insert in a 3:1 ration or a 1:1 ration for parts with the same size.
Transformation of E.Coli cells
5 μL of ligated DNA + 50 μL of chemocompetent E.coli, well mixed by resuspension with pipette Incubation on ice for 30 minutes Heat shock at 42°C for 42 seconds Incubation on ice for 5 minutes Addition of 250 μL of SOC medium Incubation at 37°C for 45 minutes Plating 150 μL of bacterial mixture on petri dish with correct antibiotics Incubation at 37°C overnight
DNA purification (“prep”)
Macherey-Nagel’s NucleoSpin® Plasmid - Isolation of high- and low-copy plasmid DNA from E. coli
Agarose-gel cleanup
Macherey-Nagel’s Gel Extraction
References
- 1. Nobbs, A. H., Jenkinson, H. F. & Jakubovics, N. S. Stick to Your Gums. J. Dent. Res. 90, 1271–1278 (2011).
- 2. Jonasson, A. et al. Innate immunity glycoprotein gp-340 variants may modulate human susceptibility to dental caries. BMC Infect. Dis. 7, 57 (2007).
- 3. Fan, M. W. et al. A DNA vaccine encoding a cell-surface protein antigen of ''Streptococcus mutans'' protects gnotobiotic rats from caries. J Dent Res 81, 784–787 (2002).
- 4. Munro, G. H. et al. A protein fragment of streptococcal cell surface antigen I / II which prevents adhesion of A Protein Fragment of Streptococcal Cell Surface Antigen I / II Which Prevents Adhesion of ''Streptococcus mutans''. 61, 4590–4598 (1993).
- 5. Vacca-Smith, A. M. & Bowen, W. H. Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva coated hydroxyapatite, and bacterial surfaces. Arch. Oral Biol. 43, 103–110 (1998).
- 6. Bowen, W. H. & Koo, H. Biology of streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86 (2011).
- 7. Kopec, L. K. & Vacca-Smith, A. M. Structural aspects of glucans formed in solution and on the surface of hydroxyapatite. Glycobiology 7, 929–934 (1997).

