(Prototype team page) |
|||
| Line 1: | Line 1: | ||
| − | |||
<html> | <html> | ||
| + | <style> | ||
| + | img[data-action="zoom"] { | ||
| + | cursor: pointer; | ||
| + | cursor: -webkit-zoom-in; | ||
| + | cursor: -moz-zoom-in; | ||
| + | } | ||
| + | .zoom-img, | ||
| + | .zoom-img-wrap { | ||
| + | position: relative; | ||
| + | z-index: 666; | ||
| + | -webkit-transition: all 300ms; | ||
| + | -o-transition: all 300ms; | ||
| + | transition: all 300ms; | ||
| + | } | ||
| + | img.zoom-img { | ||
| + | cursor: pointer; | ||
| + | cursor: -webkit-zoom-out; | ||
| + | cursor: -moz-zoom-out; | ||
| + | } | ||
| + | .zoom-overlay { | ||
| + | z-index: 420; | ||
| + | background: #fff; | ||
| + | position: fixed; | ||
| + | top: 0; | ||
| + | left: 0; | ||
| + | right: 0; | ||
| + | bottom: 0; | ||
| + | pointer-events: none; | ||
| + | filter: "alpha(opacity=0)"; | ||
| + | opacity: 0; | ||
| + | -webkit-transition: opacity 300ms; | ||
| + | -o-transition: opacity 300ms; | ||
| + | transition: opacity 300ms; | ||
| + | } | ||
| + | .zoom-overlay-open .zoom-overlay { | ||
| + | filter: "alpha(opacity=100)"; | ||
| + | opacity: 1; | ||
| + | } | ||
| + | .zoom-overlay-open, | ||
| + | .zoom-overlay-transitioning { | ||
| + | cursor: default; | ||
| + | } | ||
| + | .mdc__toolbar--fixed{top: 16px;} | ||
| + | @font-face {font-family: 'Material Icons'; font-style: normal; font-weight: 400; src: local('Material Icons'), local('MaterialIcons-Regular'), url(https://static.igem.org/mediawiki/2017/d/d5/Lanzhou_wiki_MaterialIcons-Regular.woff) format('woff'); } .material-icons {font-family: 'Material Icons'; font-weight: normal; font-style: normal; font-size: 24px; line-height: 1; letter-spacing: normal; text-transform: none; display: inline-block; white-space: nowrap; word-wrap: normal; direction: ltr; -webkit-font-feature-settings: 'liga'; -webkit-font-smoothing: antialiased; }#home_logo, #sideMenu { display:none; } #sideMenu, #top_title, .patrollink {display:none;} #content { width:100%; padding:0px; margin-top:-7px; margin-left:0px;} body {background-color:white; margin: 0;} #bodyContent h1, #bodyContent h2, #bodyContent h3, #bodyContent h4, #bodyContent h5 { margin-bottom: 0px; }#top_title{display:none; }#top_menu_under {position:relative; width:100%; height:18px } #top_menu_14 {position:fixed; width:100%; top:0px; left:0px; height:16px; background-color:#383838; border-bottom:2px solid black; z-index:9999 } #top_menu_inside {display:block; position:relative; width:960px; height:100%; margin:0px auto; padding:0 10px; border-left:1px solid #c8c8c8; border-right:1px solid #c8c8c8; font-family:'arial',sans-serif; font-size:12px; font-weight:400; color:#ffffff } #top_menu_inside ul {list-style:none; margin:0; padding:0 } #top_menu_inside li {float:left; padding:0px 25px 0px 20px; cursor:pointer; margin:0 } #top_menu_inside .has_submenu:hover {background-color:#000000; color:white; text-decoration:none } #top_menu_inside #user_item {float:right; padding-top:2px; margin-right:100px; cursor:pointer; color:orange } #top_menu_inside #user_item:hover {background-color:#000000; color:#ffc864 } #top_menu_inside #bars_item {position:absolute; top:1px; left:960px; z-index:2; cursor:pointer } #top_menu_inside #bars_box {position:absolute; right:0px; top:17px; min-height:100px; min-width:100px; color:black; background-color:white; border:1px solid black; box-shadow:rgba(50,50,50,0.75) 1px 1px 10px 0px; z-index:1 } #top_menu_inside .submenu {display:none; position:absolute; z-index:10; top:16px; left:20px; width:900px; min-height:80px; background-color:#000000; padding:10px 15px 20px 15px; box-shadow:rgba(10,10,10,0.75) 0px 5px 15px 0px; color:#ffffff } #top_menu_inside .section {float:left; width:190px; font-family:helvitica,sans-serif; font-style:normal; font-weight:400; font-size:14px; line-height:22px; text-align:justify } #top_menu_inside .section .google span {padding-left:20px } #top_menu_inside .section .google span:hover,#top_menu_inside .section #page_go:hover {text-decoration:none; color:orange } #top_menu_inside .h1 {font-style:normal; font-size:18px; line-height:24px; text-weight:600; text-decoration:underline; text-align:left } #top_menu_inside .intro {color:#ffffff; font-style:italic; font-weight:normal; font-size:16px } #top_menu_inside .has_submenu .intro a {color:#ffffff; cursor:pointer } #top_menu_inside .has_submenu .intro a:hover {color:orange } | ||
| + | .mdc-toolbar--fixed{top: 16px !important;}</style> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <script type="text/javascript" src="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/Javascript/jquery1&action=raw&ctype=text/javascript"></script> | ||
| − | < | + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS&action=raw&ctype=text/css"> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/bookstrap&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/demo&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/jquery_DonutWidget&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/jquerysctipttop&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/linearicons&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/magnific_popup&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/nice_select&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/owl_carousel&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/templatemo_style&action=raw&ctype=text/css"> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/CSS/main&action=raw&ctype=text/css"> | ||
| + | <div class="bg-wetlab"> | ||
| + | <!-- Start banner Area --> | ||
| + | <section class="generic-banner-safety relative"> | ||
| + | <!-- Start Header Area --> | ||
| + | <header class="default-header"> | ||
| + | <nav class="navbar navbar-expand-lg navbar-light"> | ||
| + | <div class="container"> | ||
| + | <a class="navbar-brand" href="Home.html" style="padding: 0px;"> | ||
| + | <img src="img/T--LZU-CHINA--logo.png" alt="" height="50"> | ||
| + | </a> | ||
| + | <button class="navbar-toggler" type="button" data-toggle="collapse" data-target="#navbarSupportedContent" aria-controls="navbarSupportedContent" aria-expanded="false" aria-label="Toggle navigation"> | ||
| + | <span class="text-white lnr lnr-menu"></span> | ||
| + | </button> | ||
| + | <div class="collapse navbar-collapse justify-content-end align-items-center" id="navbarSupportedContent"> | ||
| + | <ul class="navbar-nav"> | ||
| + | <li><a href="Home.html">Home</a></li> | ||
| + | <li class="dropdown"> | ||
| + | <a class="dropdown-toggle" href="#" id="1" data-toggle="dropdown">Team</a> | ||
| + | <div class="dropdown-menu"> | ||
| + | <a class="dropdown-item" href="generic.html">Team</a> | ||
| + | <a class="dropdown-item" href="elements.html">Attribution</a> | ||
| + | <a class="dropdown-item" href="elements.html">collaborations</a> | ||
| + | </div> | ||
| + | </li> | ||
| + | <li class="dropdown"> | ||
| + | <a class="dropdown-toggle" href="Project.html" id="2" data-toggle="dropdown">Project</a> | ||
| + | <div class="dropdown-menu"> | ||
| + | <a class="dropdown-item" href="Project.html">Description</a> | ||
| + | <a class="dropdown-item" href="generic.html">Design</a> | ||
| + | <a class="dropdown-item" href="elements.html">part</a> | ||
| + | <a class="dropdown-item" href="generic.html">experiment</a> | ||
| + | <a class="dropdown-item" href="Safety.html">safety</a> | ||
| + | </div> | ||
| + | </li> | ||
| + | <li class="dropdown"> | ||
| + | <a class="dropdown-toggle" href="#" id="3" data-toggle="dropdown">wet lab</a> | ||
| + | <div class="dropdown-menu"> | ||
| + | <a class="dropdown-item" href="generic.html">result</a> | ||
| + | <a class="dropdown-item" href="elements.html">future work</a> | ||
| + | <a class="dropdown-item" href="elements.html">interlab</a> | ||
| + | <a class="dropdown-item" href="elements.html">notebook</a> | ||
| + | </div> | ||
| + | </li> | ||
| + | <li><a href="#secvice">dry lab</a></li> | ||
| + | <li class="dropdown"> | ||
| + | <a class="dropdown-toggle" href="#" id="4" data-toggle="dropdown">HP</a> | ||
| + | <div class="dropdown-menu"> | ||
| + | <a class="dropdown-item" href="generic.html">intergrated hp</a> | ||
| + | <a class="dropdown-item" href="elements.html">public engagement</a> | ||
| + | </div> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </div> | ||
| + | </nav> | ||
| + | </header> | ||
| + | <!-- End Header Area --> | ||
| + | <div class="container" style="height: 100%"> | ||
| + | <div class="row height align-items-center justify-content-center" style="height: 90%"> | ||
| + | <div class="col-lg-10"> | ||
| + | <div class="generic-banner-content"> | ||
| + | <h2 class="text-white">Result</h2> | ||
| − | <div | + | </div> |
| − | + | </div> | |
| − | < | + | </div> |
| − | + | </div> | |
| − | + | </section> | |
| − | < | + | <!-- End banner Area --> |
| − | < | + | |
| − | < | + | <!-- About Generic Start --> |
| + | <div class="main-wrapper"> | ||
| + | <div class="location" > | ||
| + | <p style="text-align: left;"><a href="#p1" style="font-size: 14px;color: #FFFFFF;margin-bottom: 16px;">1.Overview</a></p> | ||
| + | <p style="text-align: left;"><a href="#p2" style="font-size: 14px;color: #FFFFFF;margin-bottom: 16px;">2.Parts and plasmid construct</a></p> | ||
| + | <p style="text-align: left;"><a href="#p3" style="font-size: 14px;color: #FFFFFF;margin-bottom: 20px;">3.Exosome-booster stable cell line construction & function </a></p> | ||
| + | <p style="text-align: left;"><a href="#p4" style="font-size: 14px;color: #FFFFFF;margin-bottom: 20px;">4.miR attacker stable cell line construction and function verification</a></p> | ||
| + | <p style="text-align: left;"><a href="#p5" style="font-size: 14px;color: #FFFFFF;margin-bottom: 20px;">5.Functional verification of the induced promoter</a></p> | ||
| + | <p style="text-align: left;"><a href="#p6" style="font-size: 14px;color: #FFFFFF;margin-bottom: 20px;">6.Bioinformatics data of three miRNAs</a></p> | ||
| + | <p class="scroll_top" style="text-align: center;font-size: 16px;">RETURN TOP</p> | ||
| + | </div> | ||
| + | <!-- Start Generic Area --> | ||
| + | <section class="about-generic-area pt-100" id="p1"> | ||
| + | <div class="container" style="background: rgb(255,255,255)"> | ||
| + | <div style="height: 50px"></div> | ||
| + | <p style="font-size:20px;font-family: 'Segoe Script';color: darkred">On this page you can find out all the experiments result, including the results tested in the lab and the results obtained by bioinformatics analysis | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/2/24/T--LZU-CHINA--experiment.jpg" class="img-fluid mr-20 mb-20"style="width: 95%" ></div> | ||
| + | <div style="height: 20px" id="p2"></div> | ||
| + | <h3 class="about-title mb-30">Parts and plasmid construct</h3> | ||
| + | <div class="col-md-12"> | ||
| + | <h6 style="color: black">1. Gel electrophoresis experiments</h6> | ||
| + | <p class="para2" >Here are some of the agarose gel electrophoresis results of our experiments when we construct parts:</p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/e/e5/T--LZU-CHINA--result1.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2" >Figure1. Three genes of exosome booster (hSDC4, Steap3, nadB) were detected by agarose gel electrophoresis. These three genes could help to enhance cell to secrete exosome.</p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/3/3c/T--LZU-CHINA--result2.png" class="img-fluid mr-20 mb-20"style="width: 25%" ></div> | ||
| + | <p class="para2" >Figure2. Here is a gel electrophoresis of three microRNAs we used as attacker to inhibit gastric cancer cell (miR-135b-3p; miR769-5p; miR-942-5p )</p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/9/97/T--LZU-CHINA--result3.png" class="img-fluid mr-20 mb-20"style="width: 50%" ></div> | ||
| + | <p class="para2" >Figure3. Four inducible regulatory sequences (Biotin induced system: BirA-VP16; Hypoxia induction system: pHRE-miniCMV; Galactose inducing system: pGAL1; Tetracycline induction system: pTRE) are used to regulate microRNA and exsome booster expression.</p> | ||
| + | <h6 style="color: black">2.Clones and plate</h6> | ||
| + | <p class="para2" >We transformed parts connected to pSB1C3 into E.coli DH5alpha, spread in chloramphenicol tablets and cultured it overnight under 37℃.</p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/0/03/T--LZU-CHINA--result4.png" class="img-fluid mr-20 mb-20"style="width: 30%" ></div> | ||
| + | <p class="para2" >Figure4. It is a typical figure that bacterial clones were grown on chloramphenicol plate. | ||
| + | </p> | ||
| + | </div> | ||
| − | <div class=" | + | <div style="height: 20px" id="p3"></div> |
| + | <h3 class="about-title mb-30" >Exosome-booster stable cell line construction & function </h3> | ||
| + | <div class="col-md-12"> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/b/b4/T--LZU-CHINA--wet1.png" class="img-fluid mr-20 mb-20"style="width: 60%" ></div> | ||
| + | <h6 style="color: black">1. Exosome-booster stable cell line construction</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/8/8d/T--LZU-CHINA--result5.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure5. PCDH plasmid with exosome booster and CopGFP reporting system was transfected into HEK293T cells stably, and fluorescence before and after transfection was detected by inverted fluorescence microscopy. Figure A is the HEK293T cells of the untransformed plasmid, while figure B is the HEK293T cells of the transformed exosome booster. It can be observed that transfected cells can be stimulated to produce intense fluorescence. Figure. C and D were the fluorescence images of the control group at 100 times (figure. C) and the experimental group (figure. D). | ||
| + | </p> | ||
| + | <h6 style="color: black">2. Transmission electron microscopy (TEM) detection</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/0/07/T--LZU-CHINA--result6.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure6. We extracted exosomes from HEK293T cell and negatively stained it with phosphotungstic acid. Subsequently, we use TEM to detect it in 100nm. The experimental results demonstrated that HEK293T expressed exosome booster genes (figure B) could dramatically increase its exosome secretion compare with control cell (figure A) | ||
| + | </p> | ||
| + | <h6 style="color: black">3. NTA analysis</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/7/7b/T--LZU-CHINA--resultNTA.png" class="img-fluid mr-20 mb-20"style="width: 50%" ></div> | ||
| + | <p class="para2">Figure7. The NTA analysis refers to the nanoparticle tracking analysis. The experimental results showed that HEK293T cell overexpressing exosome booster had significantly higher granules numbers at 100nm and 250nm than the control cell (about 23 times as much as the control cell). This indicates that exosome booster can increase the number of exosomes secreted by 293T cell. | ||
| + | </p> | ||
| + | <h6 style="color: black">4. Western Blotting to validate the expression of exosome booster genes</h6> | ||
| + | <p class="para2">Protein sample extracted from engineering HEK293T cell was divided into two parts. One was used for staining to verify the presence of protein.</p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/b/bf/T--LZU-CHINA--result7.png" class="img-fluid mr-20 mb-20"style="width: 30%" ></div> | ||
| + | <p class="para2">Figure8. The staining results indicated that the protein was extracted from engineering HEK293T cell. Another sample was used to do wester blotting to validate whether the exosome booster gene express or no. | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/f/fd/T--LZU-CHINA--result8.png" class="img-fluid mr-20 mb-20"style="width: 50%" ></div> | ||
| + | <p class="para2">Figure9. The WB result showed that all three exosome booster genes were successfully expressed in HEK293T. Because nadB is derived from bacteria, there's no corresponding antibody. We added 6 histidine at the 3 'end of the nadB ‘s coding site and used histidine antibodies for its detection. | ||
| + | </p> | ||
| + | <h6 style="color: black">5. Confocal microscopy analysis of exosome uptake experiments</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/b/b3/T--LZU-CHINA--result9.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure10.To detect whether exosomes can enter gastric cancer cells, we used confocal microscopy to observe the uptake of gastric cancer cells MKN45 and SGC7901 exosomes. First, exosomes were extracted from 293T cells and stained with PKH26 exosomes (red fluorescent dye). The MKN45 and SGC7901 cells were stained with a FITC dye. Exosomes and gastric cancer cells were incubated together for 6 hours. Subsequently, the uptake of exosomes by gastric cancer cells was examined by fluorescence confocal microscopy. Figure. A and C indicate that gastric cancer cells MKN45 and SGC7901 can ingested exosomes. Figure. B and D showed the cytoskeletal structures of MKN45 and SGC7901 in gastric cancer cells detected by β-tubulin antibody. | ||
| + | </p> | ||
| + | </div> | ||
| + | <div style="height: 20px" id="p4"></div> | ||
| + | <h3 class="about-title mb-30" >miR attacker stable cell line construction and function verification</h3> | ||
| + | <div class="col-md-12"> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/2/23/T--LZU-CHINA--wet2.png" class="img-fluid mr-20 mb-20"style="width: 20%;" ></div> | ||
| + | <h6 style="color: black">1. The transfection effect of miR overexpressing plasmid was detected by florescence</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/d/da/T--LZU-CHINA--result10.png" class="img-fluid mr-20 mb-20"style="width: 80%;" ></div> | ||
| + | <p class="para2">Figure11. mir135b-3p overexpression vector was transfected into human embryonic renal epithelial cell line HEK293T, gastric cancer cell line MKN45 and SGC7901 through lentivirus. The cells were transfected by puromycin screening. Figure A-C shows the transfection efficiency through the mRFP reporting system under an inverted fluorescence microscope at 40 times objective lens. The results showed that mir135b-3p expression plasmid was successfully transfected into cells and expressed. Figure D-F is the observation result under 100 times objective lens. | ||
| + | </p> | ||
| + | <h6 style="color: black">2. Using cell wound scratch assay to analyze cell migration</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/8/8b/T--LZU-CHINA--result11.png" class="img-fluid mr-20 mb-20"style="width: 80%;" ></div> | ||
| + | <p class="para2">Figure12. We examined the effects of mir-135b-3p on gastric cancer cell lines MKN45 and SGC7901 through scratch test. It can be observed from the experimental results that the control gastric cancer cells almost healed after 24 hours of scratch, while mir-135b-3p significantly inhibited the migration ability of gastric cancer cell lines MKN45 and SGC7901. | ||
| + | </p> | ||
| + | <h6 style="color: black">3. Using CCK8 (Cell Counting Kit-8) experiment to detect cell proliferation.</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/d/d4/T--LZU-CHINA--result12.png" class="img-fluid mr-20 mb-20"style="width: 40%;" ></div> | ||
| + | <p class="para2">Figure13. Because the cck-8 reagent can be reduced by dehydrogenase in the cell's mitochondria to form a highly water-soluble yellow formazan. The amount of formazan is directly proportional to the number of living cells. Therefore, we used cck-8 to detect the number of living cells. Because 450 nm is specific absorption peak of formazan, we used Abs 450 to detect formazan. The results showed that overexpression of miR-135b-3p could significantly reduce the number of gastric cancer cell MKN45. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/3/31/T--LZU-CHINA--result13.png" class="img-fluid mr-20 mb-20"style="width: 40%;" ></div> | ||
| + | <p class="para2">Figure14. As shown in the previous experiment, overexpression of miR-135b-3p significantly reduced the number of gastric cancer cell SGC7901. The decline in the number of cells on the fifth day may be due to the depletion of nutrients in the cell culture medium. | ||
| + | </p> | ||
| + | <h6 style="color: black">4. Using transwell experiment to analyze cell invasion ability.</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/0/08/T--LZU-CHINA--result14.png" class="img-fluid mr-20 mb-20"style="width: 40%;" ></div> | ||
| + | <p class="para2">Figure15. The tanswell chamber is divided into two parts by polycarbonate film and matrix glue. Tumor cells were added to the upper compartment and culture medium was added to the lower compartment. Due to the high nutrient content of the lower compartment, tumor cells will pass through the matrix glue and migrate down the chamber. The results showed that overexpression of mir-135b-3p inhibited the invasion of gastric cancer cell MKN45. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/3/3a/T--LZU-CHINA--result15.png" class="img-fluid mr-20 mb-20"style="width: 40%;" ></div> | ||
| + | <p class="para2">Figure16. Cancer cells need to secrete protease to digest the stromal glue before they can pass through the compartment. We measured the number of cells in the lower chamber using the cck-8 reagent. The results show that overexpression of mir-135b-3p inhibited the invasion of gastric cancer cell SGC7901. | ||
| + | </p> | ||
| + | <h6 style="color: black">5. Flow cytometry(FCM) shows cell cycle was inhibited by miR-135b-3p</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/1/1c/T--LZU-CHINA--result16.png" class="img-fluid mr-20 mb-20"style="width: 60%;" ></div> | ||
| + | <p class="para2">Figure17. The effect of overexpression of miR-135b-3p on the cell cycle of gastric cancer cell line MKN45 was analyzed by using flow cytometry. A complete cell cycle includes the M phase (cell division phase) and the interphase. Interphase is divided into phase G1 (early DNA synthesis), S (DNA synthesis) and G2 (late DNA synthesis). As we can be observed from this figure, the number of cells in the G1 phase of MKN45 cells overexpressing miR-135b-3p increased, while the number of cells in the G2 phase decreased. This indicated that mir-135b-3p affected the DNA synthesis of gastric cancer cell MKN45, i.e. S phase. | ||
| + | </p> | ||
| + | </div> | ||
| + | <div style="height: 20px" id="p5"></div> | ||
| + | <h3 class="about-title mb-30" >Functional verification of the induced promoter</h3> | ||
| + | <div class="col-md-12"> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/1/11/T--LZU-CHINA--wet3.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/8/89/T--LZU-CHINA--wet4.png" class="img-fluid mr-20 mb-20"style="width: 40%" ></div> | ||
| + | <p class="para2">To be able to regulate our exosome enhancement genes and expression genes. We designed different control systems. The hypoxia-inducible regulatory system regulates three exosomes. The biotin-inducing system regulates the expression of mir135b-3p. The tetracycline induction system regulates mir-942-5p.The galactose regulatory system regulates mir-769-5p.Hypoxia induction system was constructed in HRE293T and MGC803 cells, and biological induction regulation system was constructed in 293T cells. | ||
| + | </p> | ||
| + | <h6 style="color: black">1. Detection of pHRE promoter</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/d/d2/T--LZU-CHINA--result17.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure18. We tested the induced expression of pHRE (5 reapts) with CopGFP. CoCl(II) is used to create anoxic conditions. The effect of pHRE in HEK 293T cells was detected in the left image, and the experimental results showed that the fluorescence intensity of CopGFP gradually increased with the increase of CoCl(II) concentration. At the concentration of 500uM, the fluorescence intensity decreased significantly. This is the result of high concentrations of CoCl(II) inhibiting cell activity. A similar effect was observed in MGC803 cells. In addition, the fluorescence intensity of MGC803 is generally lower than that of HEK293T, which may be due to the poor expression efficiency of MGC803 as a differentiated cell. | ||
| + | </p> | ||
| + | <h6 style="color: black">2. Detection of biotin inducible system </h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/7/78/T--LZU-CHINA--result18.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure19. The function of biotin-inducing system was tested by HEK 293T. We first stably transfected the vector that expressed birA-VP16 gene driven by pEF1a promoter into HEK 293T cells. The plasmid with the O-miniCMV-mRFP element is then transferred to the engineering 293T cells. The result of the left figure is that the mRFP expression level of engineered HEK293T cells was measured at 4h under different biotin concentrations. At the same time, fluorescence intensity of mRFP was measured at different times at the concentration of 100uM biotin (right figure). From the experimental results, the mRFP fluorescence intensity gradually leveled off at when incubated for about 40 hours. | ||
| + | </p> | ||
| + | </div> | ||
| − | <div | + | <div style="height: 20px" id="p6"></div> |
| − | <h3> | + | <h3 class="about-title mb-30" >Bioinformatics data of three miRNAs</h3> |
| + | <div class="col-md-12"> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/0/02/T--LZU-CHINA--wet5.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">The effects of different microRNAs on gene expression and gene pathways were analyzed and predicted in the KEGG database. We predicted miR-135b-3p, miR-769-5p and miR-942-5p, respectively, the inhibitory ability of these three microRNAs to other genes, the gene pathways that may be affected and the interactions between these gene pathways and analyzed the interaction network between the same functional proteins that are affected. We also predicted the genetic effects of all three on human cells at the same time. These bioinformatics results suggest that this is the kind of microRNA that has complex regulatory networks. Combined with our phenotypic results (cck-8 analysis, transwell test, flow cytometry analysis, scratch test), miR-135b-3p, miR-769-5p and miR-942-5p finally affected the activity, migration and DNA synthesis of gastric cancer cells through these regulatory networks. In addition, bioinformatic analysis indicates that much work remains to be done. Many regulatory mechanisms still need to be validated and interpreted experimentally. Nevertheless, as an important regulatory device in human organism, microRNA has a promising prospect in the application of cancer treatment. | ||
| + | </p> | ||
| + | <h6 style="color: black">1.Biological information analysis of miR-135b-3p </h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/5/5f/T--LZU-CHINA--result19.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure20. According to bioinformatics prediction analysis, miR-135b-3p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-135b-3p expression was at a high level, most genes were at a low level. As the expression level of miR-135b-3p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/3/37/T--LZU-CHINA--result20.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure21. The gene pathways that miR-135b-3p may affected and the interactions between these gene pathways. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/d/db/T--LZU-CHINA--result21.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure22.The interaction network between the same functional proteins affected by miR-135b-3p. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/d/d9/T--LZU-CHINA--result22.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure23. GO (Gene Ontology) analysis. The molecular function affected by miR-135b-3p were analyzed by GO database, and there was a total of 20 molecular function related to miR-135b-3p. The color and size of the histogram represent the P value. The darker the color, the smaller the P value, the stronger the correlation between this function and miR-135b-3p. | ||
| + | </p> | ||
| + | <h6 style="color: black">2. Biological information analysis of miR-942-5p </h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/2/25/T--LZU-CHINA--result23.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure24. According to bioinformatics prediction analysis, miR-942-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-942-5p expression was at a high level, most genes were at a low level. As the expression level of miR-942-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/5/5c/T--LZU-CHINA--result24.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure25. The gene pathways that miR-942-5p may affected and the interactions between these genes. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/9/9c/T--LZU-CHINA--result25.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure26. The interaction network between the same functional proteins affected by miR-942-5p. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/4/40/T--LZU-CHINA--result26.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure27. GO (Gene Ontology) analysis. The molecular function affected by miR-942-5p were analyzed by GO database, and there was a total of 20 molecular function related to miR-942-5p. The color and size of the histogram represent the P value. The darker the color, the smaller the P value, the stronger the correlation between this function and miR-942-5p. | ||
| + | </p> | ||
| + | <h6 style="color: black">3. Biological information analysis of miR-769-5p </h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/2/27/T--LZU-CHINA--result27.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure28. According to bioinformatics prediction analysis, miR-769-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/f/f0/T--LZU-CHINA--result28.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure29. The gene pathways that miR-769-5p may affected and the interactions between these genes. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/f/fd/T--LZU-CHINA--result29.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure30. The interaction network between the same functional proteins affected by miR-769-5p. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/9/9d/T--LZU-CHINA--result30.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure31. According to bioinformatics prediction analysis, miR-769-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes. | ||
| + | </p> | ||
| + | <h6 style="color: black">4. Biological information analysis of miR-135b-3p, miR-942-5p and miR-769-5p interaction</h6> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/3/3b/T--LZU-CHINA--result31.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure32. The gene pathways that miR-135b-3p, miR-942-5p and miR-769-5p may affected and the interactions between these genes were predicted by Biological information analysis. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/1/17/T--LZU-CHINA--result32.png" class="img-fluid mr-20 mb-20"style="width: 55%" ></div> | ||
| + | <p class="para2">Figure33. The interaction network between the same functional proteins affected by miR-135b-3p, miR-942-5p and miR-769-5p. | ||
| + | </p> | ||
| + | <div class="myimg"><img src="https://static.igem.org/mediawiki/2018/c/c3/T--LZU-CHINA--result33.png" class="img-fluid mr-20 mb-20"style="width: 70%" ></div> | ||
| + | <p class="para2">Figure34. The functional molecules affected by miR-135b-3p, miR-942-5p and miR-769-5p were analyzed by GO database. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-135b-3p, miR-942-5p and miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-135b-3p, miR-942-5p and miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes. | ||
| + | </p> | ||
| − | < | + | </div> |
| + | </div> | ||
| + | </section> | ||
| + | <!-- End Generic Start --> | ||
| − | < | + | <!-- Start Generic Area --> |
| − | < | + | <section class="about-generic-area pb-100 pt-100" id="safety2"> |
| − | + | <div class="container border-top-generic" style="background: rgb(255,255,255)"> | |
| − | < | + | |
| − | </div> | + | </div> |
| + | </section> | ||
| + | <!-- End Generic Start --> | ||
| + | <!-- start footer Area --> | ||
| + | <footer class="footer-area section-gap" > | ||
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col-lg-6 col-md-6 col-sm-6"> | ||
| + | <div class="single-footer-widget"> | ||
| + | <h6>About Us</h6> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/e/ee/T--LZU-CHINA--xiaohui.png" style="height: 120px"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/d/df/T--LZU-CHINA--yiyuan.png" style="height: 120px"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/4/41/T--LZU-CHINA--shengming.png" style="height: 120px"> | ||
| + | <p> | ||
| + | We wish to extend our deepest appreciation to the Lanzhou University, The First Clinical Medical College and School of Life Science for the great support and help in this project | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-lg-6 col-md-6 col-sm-6 social-widget"> | ||
| + | <div class="single-footer-widget"> | ||
| + | <h6>Contact Us</h6> | ||
| + | <p>E-mail:752786236@qq.com 1098385458@qq.com</p> | ||
| + | <p>Address:No. 222, Tianshui South Road, chengguan district, Lanzhou City, Gansu Province,730000, P. R. China.</p> | ||
| + | <p style="font-size:20px;font-family: 'Segoe Script'">We are the walker on the long journey towards science</p> | ||
| − | < | + | <p class="footer-text">Copyright ©2018 iGEM LZU-CHINA Team All rights reserved.</p> |
| − | + | </div> | |
| − | + | </div> | |
| − | + | </div> | |
| − | < | + | </div> |
| − | + | </footer> | |
| − | < | + | <!-- End footer Area --> |
| − | < | + | |
| − | </ | + | |
| − | </ | + | |
| − | < | + | |
| + | </div> | ||
| + | <!--script src="js/vendor/jquery-2.2.4.min.js"></script> | ||
| + | <script src="js/vendor/bootstrap.min.js"></script> | ||
| + | <script src="js/jquery.ajaxchimp.min.js"></script> | ||
| + | <script src="js/jquery.sticky.js"></script> | ||
| + | <script src="js/owl.carousel.min.js"></script> | ||
| + | <script src="js/jquery.nice-select.min.js"></script> | ||
| + | <script src="js/jquery.magnific-popup.min.js"></script> | ||
| + | <script src="js/main.js"></script--> | ||
| + | </div> | ||
| + | </div> | ||
| + | <script type="text/javascript" src="https://2018.igem.org/wiki/index.php?title=Template:LZU-CHINA/Javascript/jquery1&action=raw&ctype=text/javascript"></script> | ||
| + | <script type="text/javascript"> | ||
| + | jQuery(document).ready(function($){ | ||
| + | $('.scroll_top').click(function(){$('html,body').animate({scrollTop: '0px'}, 800);}); | ||
| + | }); | ||
| + | </script> | ||
| + | </div> | ||
</html> | </html> | ||
Revision as of 05:28, 16 October 2018
3.Exosome-booster stable cell line construction & function
4.miR attacker stable cell line construction and function verification
5.Functional verification of the induced promoter
6.Bioinformatics data of three miRNAs
RETURN TOP
On this page you can find out all the experiments result, including the results tested in the lab and the results obtained by bioinformatics analysis

Parts and plasmid construct
1. Gel electrophoresis experiments
Here are some of the agarose gel electrophoresis results of our experiments when we construct parts:

Figure1. Three genes of exosome booster (hSDC4, Steap3, nadB) were detected by agarose gel electrophoresis. These three genes could help to enhance cell to secrete exosome.

Figure2. Here is a gel electrophoresis of three microRNAs we used as attacker to inhibit gastric cancer cell (miR-135b-3p; miR769-5p; miR-942-5p )

Figure3. Four inducible regulatory sequences (Biotin induced system: BirA-VP16; Hypoxia induction system: pHRE-miniCMV; Galactose inducing system: pGAL1; Tetracycline induction system: pTRE) are used to regulate microRNA and exsome booster expression.
2.Clones and plate
We transformed parts connected to pSB1C3 into E.coli DH5alpha, spread in chloramphenicol tablets and cultured it overnight under 37℃.

Figure4. It is a typical figure that bacterial clones were grown on chloramphenicol plate.
Exosome-booster stable cell line construction & function

1. Exosome-booster stable cell line construction

Figure5. PCDH plasmid with exosome booster and CopGFP reporting system was transfected into HEK293T cells stably, and fluorescence before and after transfection was detected by inverted fluorescence microscopy. Figure A is the HEK293T cells of the untransformed plasmid, while figure B is the HEK293T cells of the transformed exosome booster. It can be observed that transfected cells can be stimulated to produce intense fluorescence. Figure. C and D were the fluorescence images of the control group at 100 times (figure. C) and the experimental group (figure. D).
2. Transmission electron microscopy (TEM) detection
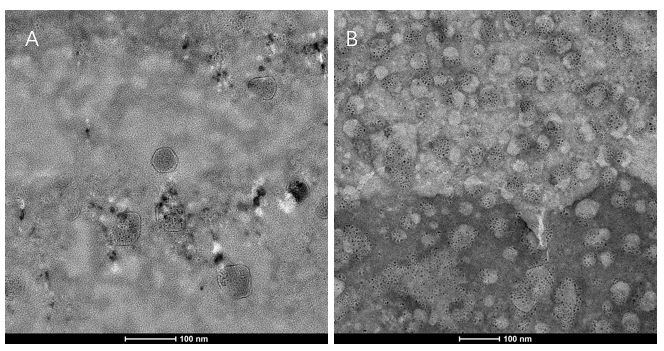
Figure6. We extracted exosomes from HEK293T cell and negatively stained it with phosphotungstic acid. Subsequently, we use TEM to detect it in 100nm. The experimental results demonstrated that HEK293T expressed exosome booster genes (figure B) could dramatically increase its exosome secretion compare with control cell (figure A)
3. NTA analysis

Figure7. The NTA analysis refers to the nanoparticle tracking analysis. The experimental results showed that HEK293T cell overexpressing exosome booster had significantly higher granules numbers at 100nm and 250nm than the control cell (about 23 times as much as the control cell). This indicates that exosome booster can increase the number of exosomes secreted by 293T cell.
4. Western Blotting to validate the expression of exosome booster genes
Protein sample extracted from engineering HEK293T cell was divided into two parts. One was used for staining to verify the presence of protein.

Figure8. The staining results indicated that the protein was extracted from engineering HEK293T cell. Another sample was used to do wester blotting to validate whether the exosome booster gene express or no.

Figure9. The WB result showed that all three exosome booster genes were successfully expressed in HEK293T. Because nadB is derived from bacteria, there's no corresponding antibody. We added 6 histidine at the 3 'end of the nadB ‘s coding site and used histidine antibodies for its detection.
5. Confocal microscopy analysis of exosome uptake experiments

Figure10.To detect whether exosomes can enter gastric cancer cells, we used confocal microscopy to observe the uptake of gastric cancer cells MKN45 and SGC7901 exosomes. First, exosomes were extracted from 293T cells and stained with PKH26 exosomes (red fluorescent dye). The MKN45 and SGC7901 cells were stained with a FITC dye. Exosomes and gastric cancer cells were incubated together for 6 hours. Subsequently, the uptake of exosomes by gastric cancer cells was examined by fluorescence confocal microscopy. Figure. A and C indicate that gastric cancer cells MKN45 and SGC7901 can ingested exosomes. Figure. B and D showed the cytoskeletal structures of MKN45 and SGC7901 in gastric cancer cells detected by β-tubulin antibody.
miR attacker stable cell line construction and function verification

1. The transfection effect of miR overexpressing plasmid was detected by florescence

Figure11. mir135b-3p overexpression vector was transfected into human embryonic renal epithelial cell line HEK293T, gastric cancer cell line MKN45 and SGC7901 through lentivirus. The cells were transfected by puromycin screening. Figure A-C shows the transfection efficiency through the mRFP reporting system under an inverted fluorescence microscope at 40 times objective lens. The results showed that mir135b-3p expression plasmid was successfully transfected into cells and expressed. Figure D-F is the observation result under 100 times objective lens.
2. Using cell wound scratch assay to analyze cell migration

Figure12. We examined the effects of mir-135b-3p on gastric cancer cell lines MKN45 and SGC7901 through scratch test. It can be observed from the experimental results that the control gastric cancer cells almost healed after 24 hours of scratch, while mir-135b-3p significantly inhibited the migration ability of gastric cancer cell lines MKN45 and SGC7901.
3. Using CCK8 (Cell Counting Kit-8) experiment to detect cell proliferation.

Figure13. Because the cck-8 reagent can be reduced by dehydrogenase in the cell's mitochondria to form a highly water-soluble yellow formazan. The amount of formazan is directly proportional to the number of living cells. Therefore, we used cck-8 to detect the number of living cells. Because 450 nm is specific absorption peak of formazan, we used Abs 450 to detect formazan. The results showed that overexpression of miR-135b-3p could significantly reduce the number of gastric cancer cell MKN45.

Figure14. As shown in the previous experiment, overexpression of miR-135b-3p significantly reduced the number of gastric cancer cell SGC7901. The decline in the number of cells on the fifth day may be due to the depletion of nutrients in the cell culture medium.
4. Using transwell experiment to analyze cell invasion ability.

Figure15. The tanswell chamber is divided into two parts by polycarbonate film and matrix glue. Tumor cells were added to the upper compartment and culture medium was added to the lower compartment. Due to the high nutrient content of the lower compartment, tumor cells will pass through the matrix glue and migrate down the chamber. The results showed that overexpression of mir-135b-3p inhibited the invasion of gastric cancer cell MKN45.

Figure16. Cancer cells need to secrete protease to digest the stromal glue before they can pass through the compartment. We measured the number of cells in the lower chamber using the cck-8 reagent. The results show that overexpression of mir-135b-3p inhibited the invasion of gastric cancer cell SGC7901.
5. Flow cytometry(FCM) shows cell cycle was inhibited by miR-135b-3p

Figure17. The effect of overexpression of miR-135b-3p on the cell cycle of gastric cancer cell line MKN45 was analyzed by using flow cytometry. A complete cell cycle includes the M phase (cell division phase) and the interphase. Interphase is divided into phase G1 (early DNA synthesis), S (DNA synthesis) and G2 (late DNA synthesis). As we can be observed from this figure, the number of cells in the G1 phase of MKN45 cells overexpressing miR-135b-3p increased, while the number of cells in the G2 phase decreased. This indicated that mir-135b-3p affected the DNA synthesis of gastric cancer cell MKN45, i.e. S phase.
Functional verification of the induced promoter


To be able to regulate our exosome enhancement genes and expression genes. We designed different control systems. The hypoxia-inducible regulatory system regulates three exosomes. The biotin-inducing system regulates the expression of mir135b-3p. The tetracycline induction system regulates mir-942-5p.The galactose regulatory system regulates mir-769-5p.Hypoxia induction system was constructed in HRE293T and MGC803 cells, and biological induction regulation system was constructed in 293T cells.
1. Detection of pHRE promoter

Figure18. We tested the induced expression of pHRE (5 reapts) with CopGFP. CoCl(II) is used to create anoxic conditions. The effect of pHRE in HEK 293T cells was detected in the left image, and the experimental results showed that the fluorescence intensity of CopGFP gradually increased with the increase of CoCl(II) concentration. At the concentration of 500uM, the fluorescence intensity decreased significantly. This is the result of high concentrations of CoCl(II) inhibiting cell activity. A similar effect was observed in MGC803 cells. In addition, the fluorescence intensity of MGC803 is generally lower than that of HEK293T, which may be due to the poor expression efficiency of MGC803 as a differentiated cell.
2. Detection of biotin inducible system

Figure19. The function of biotin-inducing system was tested by HEK 293T. We first stably transfected the vector that expressed birA-VP16 gene driven by pEF1a promoter into HEK 293T cells. The plasmid with the O-miniCMV-mRFP element is then transferred to the engineering 293T cells. The result of the left figure is that the mRFP expression level of engineered HEK293T cells was measured at 4h under different biotin concentrations. At the same time, fluorescence intensity of mRFP was measured at different times at the concentration of 100uM biotin (right figure). From the experimental results, the mRFP fluorescence intensity gradually leveled off at when incubated for about 40 hours.
Bioinformatics data of three miRNAs

The effects of different microRNAs on gene expression and gene pathways were analyzed and predicted in the KEGG database. We predicted miR-135b-3p, miR-769-5p and miR-942-5p, respectively, the inhibitory ability of these three microRNAs to other genes, the gene pathways that may be affected and the interactions between these gene pathways and analyzed the interaction network between the same functional proteins that are affected. We also predicted the genetic effects of all three on human cells at the same time. These bioinformatics results suggest that this is the kind of microRNA that has complex regulatory networks. Combined with our phenotypic results (cck-8 analysis, transwell test, flow cytometry analysis, scratch test), miR-135b-3p, miR-769-5p and miR-942-5p finally affected the activity, migration and DNA synthesis of gastric cancer cells through these regulatory networks. In addition, bioinformatic analysis indicates that much work remains to be done. Many regulatory mechanisms still need to be validated and interpreted experimentally. Nevertheless, as an important regulatory device in human organism, microRNA has a promising prospect in the application of cancer treatment.
1.Biological information analysis of miR-135b-3p

Figure20. According to bioinformatics prediction analysis, miR-135b-3p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-135b-3p expression was at a high level, most genes were at a low level. As the expression level of miR-135b-3p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes.

Figure21. The gene pathways that miR-135b-3p may affected and the interactions between these gene pathways.

Figure22.The interaction network between the same functional proteins affected by miR-135b-3p.

Figure23. GO (Gene Ontology) analysis. The molecular function affected by miR-135b-3p were analyzed by GO database, and there was a total of 20 molecular function related to miR-135b-3p. The color and size of the histogram represent the P value. The darker the color, the smaller the P value, the stronger the correlation between this function and miR-135b-3p.
2. Biological information analysis of miR-942-5p

Figure24. According to bioinformatics prediction analysis, miR-942-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-942-5p expression was at a high level, most genes were at a low level. As the expression level of miR-942-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes.

Figure25. The gene pathways that miR-942-5p may affected and the interactions between these genes.

Figure26. The interaction network between the same functional proteins affected by miR-942-5p.

Figure27. GO (Gene Ontology) analysis. The molecular function affected by miR-942-5p were analyzed by GO database, and there was a total of 20 molecular function related to miR-942-5p. The color and size of the histogram represent the P value. The darker the color, the smaller the P value, the stronger the correlation between this function and miR-942-5p.
3. Biological information analysis of miR-769-5p

Figure28. According to bioinformatics prediction analysis, miR-769-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes.

Figure29. The gene pathways that miR-769-5p may affected and the interactions between these genes.

Figure30. The interaction network between the same functional proteins affected by miR-769-5p.

Figure31. According to bioinformatics prediction analysis, miR-769-5p can inhibit 50 genes expression. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes.
4. Biological information analysis of miR-135b-3p, miR-942-5p and miR-769-5p interaction

Figure32. The gene pathways that miR-135b-3p, miR-942-5p and miR-769-5p may affected and the interactions between these genes were predicted by Biological information analysis.

Figure33. The interaction network between the same functional proteins affected by miR-135b-3p, miR-942-5p and miR-769-5p.

Figure34. The functional molecules affected by miR-135b-3p, miR-942-5p and miR-769-5p were analyzed by GO database. The redder the color, the higher the gene expression, and the bluer the color, the lower the gene expression. As you can see from the diagram. When miR-135b-3p, miR-942-5p and miR-769-5p expression was at a high level, most genes were at a low level. As the expression level of miR-135b-3p, miR-942-5p and miR-769-5p gradually decreased, most genes showed high expression level. However, some gene expression levels are not linearly correlated, which may be due to the interaction between genes.
