ElianDupre (Talk | contribs) |
ElianDupre (Talk | contribs) |
||
| Line 118: | Line 118: | ||
These are our list of results that we have successfully acquired through our two-year run developing this project. Special thanks to Lisa Oberding for helping with our Gene Design, Gene Synthesis, and Transformation. | These are our list of results that we have successfully acquired through our two-year run developing this project. Special thanks to Lisa Oberding for helping with our Gene Design, Gene Synthesis, and Transformation. | ||
</p> | </p> | ||
| − | + | <br> | |
<h1 class="subtitle">Gene Design</h1> | <h1 class="subtitle">Gene Design</h1> | ||
| Line 129: | Line 129: | ||
<li>a BslA without RFP.</li> | <li>a BslA without RFP.</li> | ||
</ul> | </ul> | ||
| + | <table style="width: 50%; float: clear;" > | ||
| + | <tr><td> | ||
| + | |||
| + | <img width="50%"src="https://static.igem.org/mediawiki/2018/f/f0/T--OLS_Canmore_Canada--Construct-mCherry.jpeg"> | ||
| + | <img style="float: right;" width="50%"src="https://static.igem.org/mediawiki/2018/4/41/T--OLS_Canmore_Canada--Construct-Without.jpeg"> | ||
| + | |||
| + | </td></tr> | ||
| + | <tr><td class="imagecaptiontext">Snapshot of our constructs</td></tr> | ||
| + | </table> | ||
<br> | <br> | ||
<p>Special care was taken to ensure the sequences did not include any extra restriction enzyme sites and to design the coding regions to be expressed successfully in BOTH E. coli and B. subtilis cells. See our parts pages for further details regarding these parts.</p> | <p>Special care was taken to ensure the sequences did not include any extra restriction enzyme sites and to design the coding regions to be expressed successfully in BOTH E. coli and B. subtilis cells. See our parts pages for further details regarding these parts.</p> | ||
Latest revision as of 12:03, 17 October 2018
RESULTS
Introduction
These are our list of results that we have successfully acquired through our two-year run developing this project. Special thanks to Lisa Oberding for helping with our Gene Design, Gene Synthesis, and Transformation.
Gene Design
The 4 constructs that we have designed, with the help of our mentor Lisa Oberding and previous iGem teams (especially Tianjin 2016), include:
- a polyethylene terephthalate hydrolase (PET-ase) fused to a red fluorescent protein, (or RFP) called mCherry, which gives the protein its colour aspect,
- a hydrophobin called BslA fused to mCherry,
- a PET-ase without the RFP, and
- a BslA without RFP.


|
| Snapshot of our constructs |
Special care was taken to ensure the sequences did not include any extra restriction enzyme sites and to design the coding regions to be expressed successfully in BOTH E. coli and B. subtilis cells. See our parts pages for further details regarding these parts.
Gene Synthesis
The gene constructs we have created with the help of Lisa were synthesized by Bio Basic and shipped to us for transformation. The plasmids were ordered both in the standard pSB1C3 backbone, as well as a second “shuttle” backbone we hope to use in the future for testing on Bacillus specifically.
Transformations
Our lab tried repeatedly to transform our cells. We had success with E. coli but were unable to transform Bacillus subtilis on our own. Thank you to Amino Labs who hosted us over the summer for a second (though unsuccessful) round of attempts at Bacillus transformation. Thankfully our mentor, Lisa Oberding from Fredsense Technologies, was able to help us successfully transform both E.coli and B.subtilis cells with each of our four constructs.
These pictures are our constructs in E.coli DH5alpha. The ones on the left are imaged under regular light and the picture on the right is under UV light. The coding system Lisa used for the test is:
- E - PET-ase
- F - mCherry PET-ase
- G - BslA
- H - mCherry BslA


|
| Regular (left), UV (right) |
These pictures are our constructs in B.subtilis SCK6. The ones on the left are imaged under regular light and the picture on the right is under UV light. The coding Lisa used for our plates are:
- E - PET-ase
- PM - mCherry PET-ase
- B - BslA
- BM - BslA-mCherry


|
| Regular (left), UV (right) |
Lysis of E. coli Cells
For full lysis protocol, go to “Lysis Protocol (E.Coli)” under the Experiments page of our wiki.
We used the lysis protocol to help extract the protein from the E.Coli, as they are not excellent secreters of proteins. Our lab does not have capabilities to purify the proteins or enzymes from the lysate, but in the future, this step will be important to further test whether the constructs do adhere selectively to the plastics. We suspect the protocol was successful due to the pink/purple colouration of the lysis buffer. In our results, we can see that F2, which is PET-ase mCherry, has a slight colouration signifying the excretion of the mCherry.
 |
| Results of the Lysis Protocol, October 5th. Notice the pink coloration on "F2". |
Testing of Constructs
Our four constructs were tested on a strip of PET plastic. We predicted there will be a slight gradient of coloration, visible to the human eye, with each construct and plastic combination. We expected to see the brightest colouration with the PET-ase and mCherry with the Hydrophobin without mCherry since the hydrophobins will make it easier to adhere better. The constructs were also tested on bubble wrap, which was our control for a non-PET plastic.
Firstly, we ran the pure mCherry through the spectrophotometer to measure the wavelength where it shows the max absorbance to have a baseline target for our upcoming tests. We determined from the results the absorbance peak was 1.011 at 587nm.
 |
| Click to see full size image |
Due to late results for B. subtilis, we only got a chance to test with the transformed E. coli cell cultures. We used the spectrophotometer and ran every construct at wavelengths ranging from 557 nm to 617 nm to test different absorbances. For the full description of the procedure, see “Spectrophotometer Readings - Cell Cultures” under the protocols page.
 |
| Click to see full size image |
 |
| Click to see full size image |
 |
| Click to see full size image |
 |
| Click to see full size image |
The graphs above are our four constructs on unlysed E.coli under the spectrophotometer. We knew the mCherry was secreted by the E.coli through the absorbance at 587 nm. All constructs that had mCherry had a high absorbance at 587 nm; 0.72 for the BslA mCherry and 0.8975 for the PET-ase mCherry. When compared to the absorbance of pure mCherry at 587 nm, 1.011, we can tell it is secreting but not entirely as it is a little bit lower than 1.011.
The next step was to run the lysed cells, that we had from our lysis protocol, under the spectrophotometer with the same procedure of testing every construct through wavelengths of 557 nm to 617 nm. For the full description of this procedure, see “Spectrophotometer Readings - Lysed Supernatants” under the protocols page.
 |
| Click to see full size image |
 |
| Click to see full size image |
 |
| Click to see full size image |
 |
| Click to see full size image |
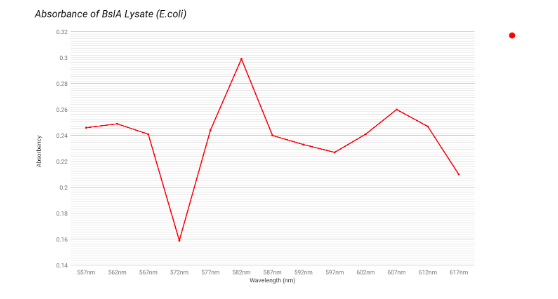
The graphs above are our four constructs on lysed E. coli under the spectrophotometer. We saw little evidence of mCherry on the lysed cells because of the low absorbance of the constructs at 587 nm. Compared to the pure mCherry’s 1.011 absorbance at 587 nm, the lysed cells with mCherry in their construct only had an absorbance range of 0.2 - 0.4, which is significantly lower. This may be due to our lack of equipment that would allow for the proper purification of our proteins. Experimenting with the proteins in their unpurified state may be preventing the expression of the mCherry. Future plans will address this deficiency.
 |
| Results for Lyse and Cells |
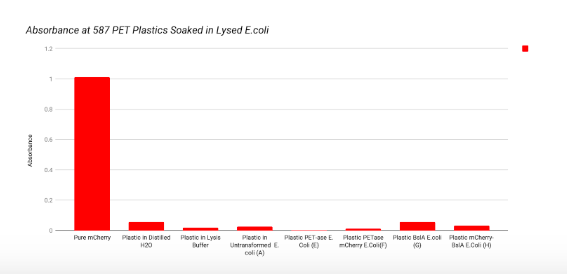
Our final step was to take strips of PET plastics and non-PET plastics and soak them in each of the constructs. See “Spectrophotometer Readings - PET and non-PET Plastic” under the “Experiments” section for the full description. After soaking the plastics we noticed a lack of colouration on the PET plastics, an indication that the mCherry did not stick to the plastics in quantities that would make them visible to the naked eye. Further testing would aim to see if soaked plastics were visible under UV light or using UV optical devices.
To test if the mCherry was still present but simply not visible to the human eye, we ran our plastics through the spectrophotometer to check absorbances at 587nm. Our results were compared with the absorbance of pure mCherry, plastic soaked in distilled water, and plastic in lysis buffer and LB broth; all at the wavelength of 587 nm. Compared to the expected maximum absorbance for mCherry, our constructs containing mCherry did not show clear signs of being successfully adhered to either plastic. Our lab is not able to purify proteins; future work will aim to collaborate with a lab that would purify our protein products and repeat these tests to check for evidence of selective binding to PET plastic. Our final results may have been affected by the plastic’s orientation on the spectrophotometer tube and it’s bendable properties. We aim to do future research to explain and prevent those properties to hopefully achieve more accurate results.
 |
| Click to see full size image |
 |
| Click to see full size image |
These two graphs show our PET and non-PET plastics that were soaked in our four constructs in transformed and untransformed E. coli. The adherence of mCherry was slightly present under the spectrophotometer through the small increase of absorbance of the PET plastics soaked in PET-ase mCherry, compared to the non-mCherry soaked PET plastics. The adherence of mCherry was visible through the spectrometer, because the PET plastic soaked in mCherry PET-ase had a higher absorbance than the PET plastic soaked in PET-ase without mCherry.
 |
| Click to see full size image |
 |
| Click to see full size image |
These last two graphs show our PET and non-PET plastics that were soaked in our four constructs in the lysed E.coli. The adherence of mCherry was slightly less present under the spectrophotometer than the transformed and untransformed E.coli. We can infer this through the small increase of absorbance of the PET plastics soaked in PET-ase mCherry, compared to the non mCherry soaked PET plastics.
Summary
We were able to successfully:
- Design 4 novel plasmid constructs
- Transform B. subtilis with all 4 constructs, successfully
- Transform E. coli with all 4 constructs, successfully
- Miniprep purified plasmids for all 4 constructs (pSB1C3 backbone only), for submission to Registry of Parts.
- Performed lysis protocols on E. coli cells
- Test our constructs on PET and non-PET plastic and run the results through the spectrometer for early quantitative results about mCherry visibility and PET adhesion
- Dried and submitted plasmid DNA for all 4 parts to the Parts Registry
Unsuccessful results:
- As expected, E. coli are not secreting protein constructs well. With lysis, we hoped to increase protein visibility, however our lab does not have protein purification capabilities at this time. This would be a logical next step for other teams with this capacity.
- Spectrophotometer results were ambiguous for many constructs - likely due to the lack of protein purification. Our spec is also very old and has limited capabilities. In future we hope to use some community lab space in our area to re-assay all constructs on a more powerful spec.
- It took a long time and many trials to get successful transformants in Bacillus. For this reason, we were unable to complete any characterization of how our parts work in the Bacillus chassis system but hope to do so in the future.

