| (5 intermediate revisions by one other user not shown) | |||
| Line 53: | Line 53: | ||
<a href="#">Overview</a> | <a href="#">Overview</a> | ||
</li> | </li> | ||
| − | <li | + | <li> |
<a href="#">Single-cell Model</a> | <a href="#">Single-cell Model</a> | ||
</li> | </li> | ||
| − | <li> | + | <li class="active"> |
<a href="#">System Analysis</a> | <a href="#">System Analysis</a> | ||
</li> | </li> | ||
| Line 259: | Line 259: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <p class="card-text">The range of strength value in the simulation is from 1e-2 to 1. The simulation time is 200000 s to guarantee that all species reach equilibrium state at last. From figure1 and figure2, we can find that the change of Vppa and GFP concentration is mainly follow the exponential function. However, the index of Vppa is positive but negative in the condition of GFP. It is reasonable because Vppa expression is promoted by promoter1 and promoter2 represses GFP expression. Considering that the small amount of GFP is difficult to be detected, we can conclude that s2 over 1e-2 is appropriate for repression of GFP expression. Figure3 shows the tendency of AHL concentration change with variable ‘s2’ and ‘s3’. It is obvious that s3 has little impact to AHL concentration but s2 plays a vital role in regulating AHL concentration. Within our parameter region, AHL concentration is much smaller than 0.1 nM, so the expression of degradation system is very low. This phenomenon can clearly be observed from figure 4. When s2 is lower than 1e-1.5 and s3 is higher than 1e-0.5, only about 5-20 M of NicA2 is generated but this concentration range is enough low to satisfy the design.</p> | + | <p class="card-text">The range of strength value in the simulation is from 1e-2 to 1. |
| + | The simulation time is 200000 s to guarantee that all species reach equilibrium | ||
| + | state at last. From figure1 and figure2, we can find that the change of Vppa and | ||
| + | GFP concentration is mainly follow the exponential function. However, the index of | ||
| + | Vppa is positive but negative in the condition of GFP. It is reasonable because | ||
| + | Vppa expression is promoted by promoter1 and promoter2 represses GFP expression. | ||
| + | Considering that the small amount of GFP is difficult to be detected, we can | ||
| + | conclude that s2 over 1e-2 is appropriate for repression of GFP expression. Figure3 | ||
| + | shows the tendency of AHL concentration change with variable ‘s2’ and ‘s3’. It is | ||
| + | obvious that s3 has little impact to AHL concentration but s2 plays a vital role in | ||
| + | regulating AHL concentration. Within our parameter region, AHL concentration is | ||
| + | much smaller than 0.1 nM, so the expression of degradation system is very low. This | ||
| + | phenomenon can clearly be observed from figure 4. When s2 is lower than 1e-1.5 and | ||
| + | s3 is higher than 1e-0.5, only about 5-20 M of NicA2 is generated but this | ||
| + | concentration range is enough low to satisfy the design.</p> | ||
</div> | </div> | ||
| Line 266: | Line 280: | ||
<h4 class="card-header">Adding nicotine</h4> | <h4 class="card-header">Adding nicotine</h4> | ||
<div class="card-body"> | <div class="card-body"> | ||
| − | <p class="card-text">From the simulation result of initial steady state, we can find that range of promoter strength from 0.1 to 1 for both promoters satisfies our requirements of low expression of GFP and NicA2. So, to simplify the simulation, we set the parameter interval of s1 and s2 to [0.1,1]. Under these settings, the difference among initial states under control of various s1/s2 combinations is not very significant. Based on this fact, we set the same initial condition for all simulation to reduce the amount and time of calculation.</p> | + | <p class="card-text">From the simulation result of initial steady state, we can find |
| + | that range of promoter strength from 0.1 to 1 for both promoters satisfies our | ||
| + | requirements of low expression of GFP and NicA2. So, to simplify the simulation, we | ||
| + | set the parameter interval of s1 and s2 to [0.1,1]. Under these settings, the | ||
| + | difference among initial states under control of various s1/s2 combinations is not | ||
| + | very significant. Based on this fact, we set the same initial condition for all | ||
| + | simulation to reduce the amount and time of calculation.</p> | ||
<div class="card-deck mb-3"> | <div class="card-deck mb-3"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/c/c8/T--USTC--Model2-Fig5.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/c/c8/T--USTC--Model2-Fig5.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 5 GFP with s2, s3, nicotine input</h4> | <h4 class="card-title">Figure 5 GFP with s2, s3, nicotine input</h4> | ||
| Line 275: | Line 296: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/4/40/T--USTC--Model2-Fig6.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/4/40/T--USTC--Model2-Fig6.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 6 GFP heat-map</h4> | <h4 class="card-title">Figure 6 GFP heat-map</h4> | ||
| Line 283: | Line 305: | ||
<div class="card-deck mb-3"> | <div class="card-deck mb-3"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/a/a7/T--USTC--Model2-Fig7.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/a/a7/T--USTC--Model2-Fig7.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 7 AHL with s2, s3, nicotine input</h4> | <h4 class="card-title">Figure 7 AHL with s2, s3, nicotine input</h4> | ||
| Line 289: | Line 312: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/c/ce/T--USTC--Model2-Fig8.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/c/ce/T--USTC--Model2-Fig8.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 8 AHL heat-map </h4> | <h4 class="card-title">Figure 8 AHL heat-map </h4> | ||
| Line 297: | Line 321: | ||
<div class="card-deck"> | <div class="card-deck"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/2/29/T--USTC--Model2-Fig9.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/2/29/T--USTC--Model2-Fig9.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 9 NicA2 with s2, s3, nicotine input</h4> | <h4 class="card-title">Figure 9 NicA2 with s2, s3, nicotine input</h4> | ||
| Line 303: | Line 328: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/d/d1/T--USTC--Model2-Fig10.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/d/d1/T--USTC--Model2-Fig10.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 10 Product with s2, s3, nicotine input</h4> | <h4 class="card-title">Figure 10 Product with s2, s3, nicotine input</h4> | ||
| Line 309: | Line 335: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <p class="card-text">We select GFP/AHLl/NicA2/3-Succinoyl-Pyridine to reflect the impact of s1/s2 to the gene circuit. The tendency of GFP and AHL change is similar that they are both sensitive to the strength of promoter2 but insensitive to the strength of promoter3. As s2 increases, both AHL and GFP expression rise to a higher level. The maximal concentration of AHL can reach 700 nM and GFP maximal concentration is about 1500 nM. This relationship shows that the signal intensity is mainly controlled by the expression level of hdnoR and the function of regulation system is strong related to the level of repression. From figure9 and figure 10, we notice that the value of NicA2 and 3-succinoyl-pyridine are in a narrow range which means that it is useless to change combinations of promoters to improve the efficiency of degradation system.</p> | + | <p class="card-text">We select GFP/AHLl/NicA2/3-Succinoyl-Pyridine to reflect the |
| + | impact of s1/s2 to the gene circuit. The tendency of GFP and AHL change is similar | ||
| + | that they are both sensitive to the strength of promoter2 but insensitive to the | ||
| + | strength of promoter3. As s2 increases, both AHL and GFP expression rise to a | ||
| + | higher level. The maximal concentration of AHL can reach 700 nM and GFP maximal | ||
| + | concentration is about 1500 nM. This relationship shows that the signal intensity | ||
| + | is mainly controlled by the expression level of hdnoR and the function of | ||
| + | regulation system is strong related to the level of repression. From figure9 and | ||
| + | figure 10, we notice that the value of NicA2 and 3-succinoyl-pyridine are in a | ||
| + | narrow range which means that it is useless to change combinations of promoters to | ||
| + | improve the efficiency of degradation system.</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 315: | Line 351: | ||
<h4 class="card-header">Conclusions about promoter strength</h4> | <h4 class="card-header">Conclusions about promoter strength</h4> | ||
<div class="card-body"> | <div class="card-body"> | ||
| − | <p class="card-text">For system optimizing, the combination of promoter1 and promoter2 is required to guarantee low expression without nicotine but relative high concentration of effective molecules. According to our simulation results, promoter1 strength within [0.1,0.3] and promter2 strength within [0.1,1] is the ideal combination for realizing the goal of design.</p> | + | <p class="card-text">For system optimizing, the combination of promoter1 and promoter2 |
| + | is required to guarantee low expression without nicotine but relative high | ||
| + | concentration of effective molecules. According to our simulation results, | ||
| + | promoter1 strength within [0.1,0.3] and promter2 strength within [0.1,1] is the | ||
| + | ideal combination for realizing the goal of design.</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 325: | Line 365: | ||
<hr> | <hr> | ||
<p class="card-text">The gene circuit can be divided to two separate plasmids:</p> | <p class="card-text">The gene circuit can be divided to two separate plasmids:</p> | ||
| − | <p class="card-text">(1) Plasmid1 contains gene Vppa, hdnoR, 6Hlno, LuxI, GFP.</p> | + | <p class="card-text">(1) Plasmid1 contains gene Vppa, hdnoR, 6Hlno, LuxI, GFP.</p> |
| − | <p class="card-text">(2) Plasmid2 contains gene LuxR, NicA2, Pnao, Sapd.</p> | + | <p class="card-text">(2) Plasmid2 contains gene LuxR, NicA2, Pnao, Sapd.</p> |
| − | <p class="card-text">These gene parts are inserted to plasmid backbone, but the copy number of plasmid varies. Copy number of genes is relevant to their expression level. This part we want to talk about the impact of plasmid copy number to the system function and choose the appropriate plasmid for experiment. We set plasmid copy number from 1 to 100 per cell. And strength of all promoters are defined to be 0.5. </p> | + | <p class="card-text">These gene parts are inserted to plasmid backbone, but the copy number of |
| + | plasmid varies. Copy number of genes is relevant to their expression level. This part we | ||
| + | want to talk about the impact of plasmid copy number to the system function and choose the | ||
| + | appropriate plasmid for experiment. We set plasmid copy number from 1 to 100 per cell. And | ||
| + | strength of all promoters are defined to be 0.5. </p> | ||
<div class="card"> | <div class="card"> | ||
<h4 class="card-header">Initial steady state</h4> | <h4 class="card-header">Initial steady state</h4> | ||
<div class="card-body"> | <div class="card-body"> | ||
| − | <p class="card-text">In the simulation process, to make sure that the system has reached steady state, we set the simulation time to 200000 s. Then we plot the change of Vppa/GFP/AHL/NicA2 concentration with respect to copy number.</p> | + | <p class="card-text">In the simulation process, to make sure that the system has |
| + | reached steady state, we set the simulation time to 200000 s. Then we plot the | ||
| + | change of Vppa/GFP/AHL/NicA2 concentration with respect to copy number.</p> | ||
<div class="card-deck mb-3"> | <div class="card-deck mb-3"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b7/T--USTC--Model2-Fig11.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b7/T--USTC--Model2-Fig11.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 11 Initial Vppa</h4> | <h4 class="card-title">Figure 11 Initial Vppa</h4> | ||
| Line 340: | Line 387: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/7/7c/T--USTC--Model2-Fig12.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/7/7c/T--USTC--Model2-Fig12.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 12 Initial GFP</h4> | <h4 class="card-title">Figure 12 Initial GFP</h4> | ||
| Line 348: | Line 396: | ||
<div class="card-deck"> | <div class="card-deck"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/8/82/T--USTC--Model2-Fig13.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/8/82/T--USTC--Model2-Fig13.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 13 Initial AHL</h4> | <h4 class="card-title">Figure 13 Initial AHL</h4> | ||
| Line 354: | Line 403: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/6/67/T--USTC--Model2-Fig14.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/6/67/T--USTC--Model2-Fig14.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 14 Initial NicA2</h4> | <h4 class="card-title">Figure 14 Initial NicA2</h4> | ||
| Line 360: | Line 410: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <p class="card-text">From figure11, the relation between plasmid1 copy number and Vppa concentration follows linear relationship. But the decline of GFP is similar to exponential function with negative index. The GFP concentration is always under 1e-5 nM with any copy number. AHL concentration is controlled by both LuxR and LuxI, so AHL change is related to the copy number of both plasmid1 and plamid2. However, figure13 shows that AHL concentration is strong relevant to plasmid1 copy number but week relevant to plasmid2 copy number. The maximal value of AHL concentration is about 7e-5 nm, 3 fold lower than AHL limit, which satisfies our expectations of initial states. The condition of NicA2 is contradicted with AHL. More plasmid2 leads to higher NicA2 quantity and the maximal concentration can reach 1.2 nM.</p> | + | <p class="card-text">From figure11, the relation between plasmid1 copy number and Vppa |
| + | concentration follows linear relationship. But the decline of GFP is similar to | ||
| + | exponential function with negative index. The GFP concentration is always under | ||
| + | 1e-5 nM with any copy number. AHL concentration is controlled by both LuxR and | ||
| + | LuxI, so AHL change is related to the copy number of both plasmid1 and plamid2. | ||
| + | However, figure13 shows that AHL concentration is strong relevant to plasmid1 copy | ||
| + | number but week relevant to plasmid2 copy number. The maximal value of AHL | ||
| + | concentration is about 7e-5 nm, 3 fold lower than AHL limit, which satisfies our | ||
| + | expectations of initial states. The condition of NicA2 is contradicted with AHL. | ||
| + | More plasmid2 leads to higher NicA2 quantity and the maximal concentration can | ||
| + | reach 1.2 nM.</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 366: | Line 426: | ||
<h4 class="card-header">Adding nicotine</h4> | <h4 class="card-header">Adding nicotine</h4> | ||
<div class="card-body"> | <div class="card-body"> | ||
| − | <h4 class="card-title">We select the initial state after 200000 s as the initial condition for simulation with nicotine. The initial concentration of extracellular nicotine is set to 0.00616 M and the simulation runs for 200000s to get complete dataset of all species change.</h4> | + | <h4 class="card-title">We select the initial state after 200000 s as the initial |
| + | condition for simulation with nicotine. The initial concentration of extracellular | ||
| + | nicotine is set to 0.00616 M and the simulation runs for 200000s to get complete | ||
| + | dataset of all species change.</h4> | ||
<div class="card-deck mb-3"> | <div class="card-deck mb-3"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/9/95/T--USTC--Model2-Fig15.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/9/95/T--USTC--Model2-Fig15.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 15 GFP with plasmid number</h4> | <h4 class="card-title">Figure 15 GFP with plasmid number</h4> | ||
| Line 375: | Line 439: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/2/2b/T--USTC--Model2-Fig16.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/2/2b/T--USTC--Model2-Fig16.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 16 AHL with plasmid number</h4> | <h4 class="card-title">Figure 16 AHL with plasmid number</h4> | ||
| Line 383: | Line 448: | ||
<div class="card-deck"> | <div class="card-deck"> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b1/T--USTC--Model2-Fig17.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b1/T--USTC--Model2-Fig17.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 17 NicA2 with plasmid number</h4> | <h4 class="card-title">Figure 17 NicA2 with plasmid number</h4> | ||
| Line 389: | Line 455: | ||
</div> | </div> | ||
<div class="card"> | <div class="card"> | ||
| − | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b6/T--USTC--Model2-Fig18.png" alt=""> | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/b/b6/T--USTC--Model2-Fig18.png" |
| + | alt=""> | ||
<div class="card-body"> | <div class="card-body"> | ||
<h4 class="card-title">Figure 18 Product with plasmid number</h4> | <h4 class="card-title">Figure 18 Product with plasmid number</h4> | ||
| Line 395: | Line 462: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <p class="card-text">We mainly analyze the change of GFP/AHL/NicA2/3-Succinoyl-Pyridine with different combinations of plamid1/plasmid2 copy number. The tendency of GFP and AHL change is very similar. Both species are sensitive to the increase of plamid1 copy number. To acquire AHL over 0.1 nm, the copy number of plasmid1 is limited to be smaller than 50 per cell. In consideration of GFP expression level, the requirements of plasmid1 copy number is to be smaller than 50 per cell as well. Figure3 and figure4 show that plasmid1 copy number over 40 per cell leads to low quantity of NicA2 and 3-Succinoyl-Pyridine. If plasmid1 copy number is smaller than 40 per cell, the NicA2 concentration rises with the increase of plamid2 copy number in a linear way. But for 3-Succinoyl-Pyridine, the decline of plasmid1 copy number and increase of plasmid2 copy number together causes more nicotine transformed to 3-Succinoyl-Pyridine.</p> | + | <p class="card-text">We mainly analyze the change of GFP/AHL/NicA2/3-Succinoyl-Pyridine |
| + | with different combinations of plamid1/plasmid2 copy number. The tendency of GFP | ||
| + | and AHL change is very similar. Both species are sensitive to the increase of | ||
| + | plamid1 copy number. To acquire AHL over 0.1 nm, the copy number of plasmid1 is | ||
| + | limited to be smaller than 50 per cell. In consideration of GFP expression level, | ||
| + | the requirements of plasmid1 copy number is to be smaller than 50 per cell as well. | ||
| + | Figure3 and figure4 show that plasmid1 copy number over 40 per cell leads to low | ||
| + | quantity of NicA2 and 3-Succinoyl-Pyridine. If plasmid1 copy number is smaller than | ||
| + | 40 per cell, the NicA2 concentration rises with the increase of plamid2 copy number | ||
| + | in a linear way. But for 3-Succinoyl-Pyridine, the decline of plasmid1 copy number | ||
| + | and increase of plasmid2 copy number together causes more nicotine transformed to | ||
| + | 3-Succinoyl-Pyridine.</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 401: | Line 479: | ||
<h4 class="card-header">Conclusions about plasmid copy number</h4> | <h4 class="card-header">Conclusions about plasmid copy number</h4> | ||
<div class="card-body"> | <div class="card-body"> | ||
| − | <p class="card-text">Under the determined combinations of promoters, it is better to choose the plasmid whose copy number is lower than 20 per cell as the backbone of plasimd1. As for plasmid2, copy number ranging from 20 to 40 per cell is an economical option for exact expression control and efficient nicotine degradation.</p> | + | <p class="card-text">Under the determined combinations of promoters, it is better to |
| + | choose the plasmid whose copy number is lower than 20 per cell as the backbone of | ||
| + | plasimd1. As for plasmid2, copy number ranging from 20 to 40 per cell is an | ||
| + | economical option for exact expression control and efficient nicotine degradation.</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 414: | Line 495: | ||
<p class="card-text">Nicotine input 0.00616 M</p> | <p class="card-text">Nicotine input 0.00616 M</p> | ||
<p class="card-text">Variable parameters:</p> | <p class="card-text">Variable parameters:</p> | ||
| + | <table class="table table-hover table-secondary"> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th>Plasmid copy number</th> | ||
| + | <th>Minimal value (per cell)</th> | ||
| + | <th>Maximal value (per cell)</th> | ||
| + | <th>Example</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td scope="row">Plasmid 1</td> | ||
| + | <td>1</td> | ||
| + | <td>20</td> | ||
| + | <td>pET28</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td scope="row">Plasmid 2</td> | ||
| + | <td>20</td> | ||
| + | <td>40</td> | ||
| + | <td>pET22b(+)</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <table class="table table-hover table-secondary"> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th>Promoter strength</th> | ||
| + | <th>Minimal value</th> | ||
| + | <th>Maximal value</th> | ||
| + | <th>Example</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td scope="row">Promoter 2 (hdnoR)</td> | ||
| + | <td>0.1</td> | ||
| + | <td>0.3</td> | ||
| + | <td>J23117</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td scope="row">Promoter 3 (LuxR)</td> | ||
| + | <td>0.1</td> | ||
| + | <td>1</td> | ||
| + | <td>J23116</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 500: | Line 581: | ||
$("body").scrollspy({ target: "#navId", offset: 50 }); | $("body").scrollspy({ target: "#navId", offset: 50 }); | ||
$("li.active a").click(); | $("li.active a").click(); | ||
| + | $("table").css("margin","0 0 16px 0"); | ||
}); | }); | ||
</script> | </script> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 21:04, 17 October 2018
System analysis
Purpose of analysis
In our project, the whole gene circuit is composed of three parts: sensing system, regulation system and degradation system. The function of sensing system is to detect the existence of nicotine and transform the chemical signal (6-Hydroxynicotine) to fluorescence signal (GFP). After receiving the chemical signal, the regulation system can activate the expression of degradation system by increasing the concentration of AHL. In this way, nicotine can be detected by the bacteria and it is economical for bacteria to spare some energy from expressing degradation enzymes to normal growth after sensing. At the same time, adequate metabolic enzymes can be synthesized for nicotine transformation. To reach the target of design, we need to insure low expression of GFP/NicA2 without nicotine, but sufficient quantity of effective or signal proteins when sensing nicotine. The level of expression is under the control of their promoter strengths and DNA copy numbers. This part we will talk about the impact of these two factors to the whole system and help the experiment design to choose the appropriate gene elements for optimizing.
Promoter strength
There are three important promoters controlling the expression and signal processing in the circuit:
(1) Promoter1 controls expression of Vppa. Vppa catalyzes the reaction from nicotine to 6-Hydroxynicotine.
(2) Promoter2 controls expression of hdnoR which binds to pHdno and represses 6Hlno/GFP/LuxI expression. 6-Hydroxynicotine is the inducer to release hdnoR repression. 6Hlno is responsible for 6-Hydroxynicotine degradation. LuxI catalyzes the reaction of AHL synthesis.
(3) Promoter3 controls expression of LuxR. A certain amount of LuxR-AHL dimer can activate the transcription of NicA22.
To describe the strength of promoter, we define variable ‘s’ as the strength of promoter. ‘s’ can be any real number in the region of [0,1]. The transcription rate of three promoters can be described as below:

Equation 1 Transcription rate and promoter strength
Initial steady state
The combination of promoter strengths need to meet the requirements of the design:
(1) Low concentration of GFP under detection limit.
(2) Low concentration of AHL to lower the basal expression of degradation system.
(3) Low concentration of NicA2 to save energy for normal growth.

Figure 1 Initial GFP expression with s1

Figure 2 Initial GFP expression with s2

Figure 3 Initial AHL expression with s2, s3

Figure 4 Initial NicA expression with s2, s3
The range of strength value in the simulation is from 1e-2 to 1. The simulation time is 200000 s to guarantee that all species reach equilibrium state at last. From figure1 and figure2, we can find that the change of Vppa and GFP concentration is mainly follow the exponential function. However, the index of Vppa is positive but negative in the condition of GFP. It is reasonable because Vppa expression is promoted by promoter1 and promoter2 represses GFP expression. Considering that the small amount of GFP is difficult to be detected, we can conclude that s2 over 1e-2 is appropriate for repression of GFP expression. Figure3 shows the tendency of AHL concentration change with variable ‘s2’ and ‘s3’. It is obvious that s3 has little impact to AHL concentration but s2 plays a vital role in regulating AHL concentration. Within our parameter region, AHL concentration is much smaller than 0.1 nM, so the expression of degradation system is very low. This phenomenon can clearly be observed from figure 4. When s2 is lower than 1e-1.5 and s3 is higher than 1e-0.5, only about 5-20 M of NicA2 is generated but this concentration range is enough low to satisfy the design.
Adding nicotine
From the simulation result of initial steady state, we can find that range of promoter strength from 0.1 to 1 for both promoters satisfies our requirements of low expression of GFP and NicA2. So, to simplify the simulation, we set the parameter interval of s1 and s2 to [0.1,1]. Under these settings, the difference among initial states under control of various s1/s2 combinations is not very significant. Based on this fact, we set the same initial condition for all simulation to reduce the amount and time of calculation.

Figure 5 GFP with s2, s3, nicotine input

Figure 6 GFP heat-map

Figure 7 AHL with s2, s3, nicotine input

Figure 8 AHL heat-map

Figure 9 NicA2 with s2, s3, nicotine input

Figure 10 Product with s2, s3, nicotine input
We select GFP/AHLl/NicA2/3-Succinoyl-Pyridine to reflect the impact of s1/s2 to the gene circuit. The tendency of GFP and AHL change is similar that they are both sensitive to the strength of promoter2 but insensitive to the strength of promoter3. As s2 increases, both AHL and GFP expression rise to a higher level. The maximal concentration of AHL can reach 700 nM and GFP maximal concentration is about 1500 nM. This relationship shows that the signal intensity is mainly controlled by the expression level of hdnoR and the function of regulation system is strong related to the level of repression. From figure9 and figure 10, we notice that the value of NicA2 and 3-succinoyl-pyridine are in a narrow range which means that it is useless to change combinations of promoters to improve the efficiency of degradation system.
Conclusions about promoter strength
For system optimizing, the combination of promoter1 and promoter2 is required to guarantee low expression without nicotine but relative high concentration of effective molecules. According to our simulation results, promoter1 strength within [0.1,0.3] and promter2 strength within [0.1,1] is the ideal combination for realizing the goal of design.
Plasmid copy number
The gene circuit can be divided to two separate plasmids:
(1) Plasmid1 contains gene Vppa, hdnoR, 6Hlno, LuxI, GFP.
(2) Plasmid2 contains gene LuxR, NicA2, Pnao, Sapd.
These gene parts are inserted to plasmid backbone, but the copy number of plasmid varies. Copy number of genes is relevant to their expression level. This part we want to talk about the impact of plasmid copy number to the system function and choose the appropriate plasmid for experiment. We set plasmid copy number from 1 to 100 per cell. And strength of all promoters are defined to be 0.5.
Initial steady state
In the simulation process, to make sure that the system has reached steady state, we set the simulation time to 200000 s. Then we plot the change of Vppa/GFP/AHL/NicA2 concentration with respect to copy number.
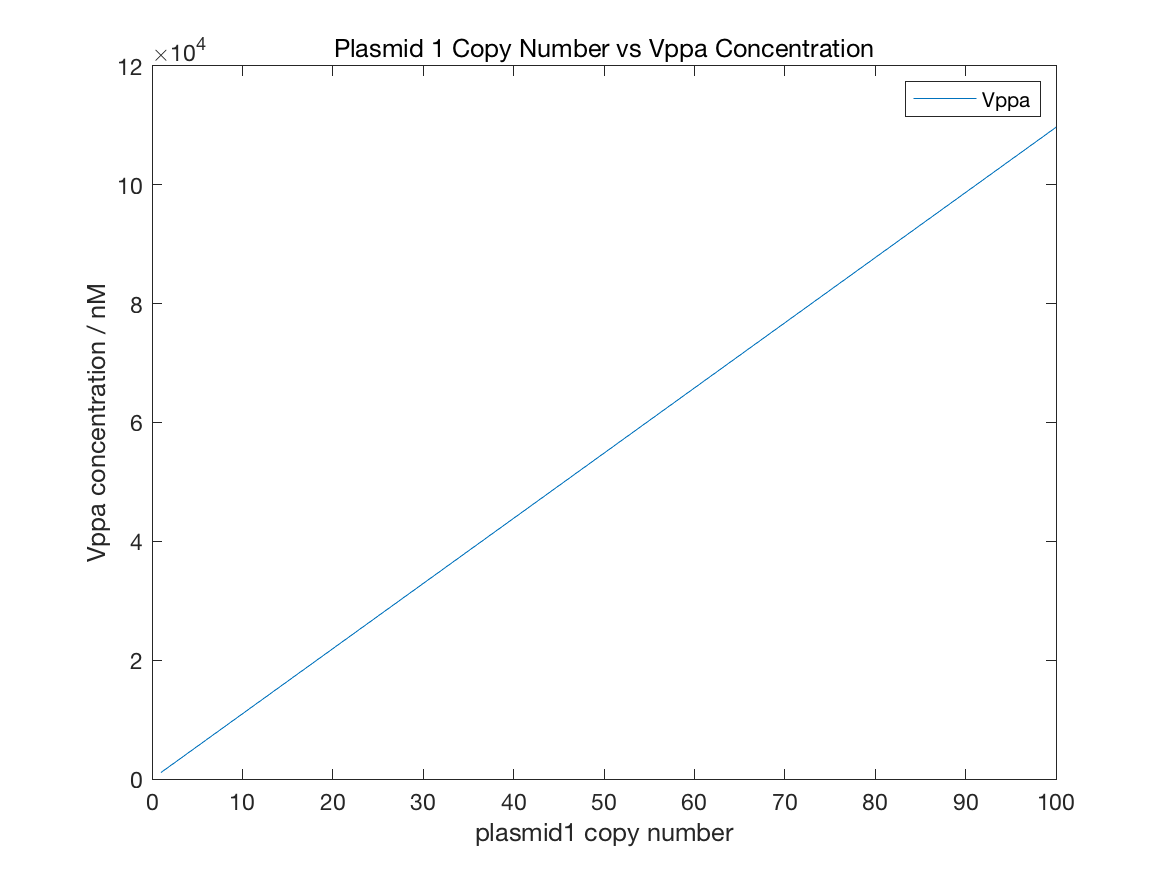
Figure 11 Initial Vppa

Figure 12 Initial GFP

Figure 13 Initial AHL

Figure 14 Initial NicA2
From figure11, the relation between plasmid1 copy number and Vppa concentration follows linear relationship. But the decline of GFP is similar to exponential function with negative index. The GFP concentration is always under 1e-5 nM with any copy number. AHL concentration is controlled by both LuxR and LuxI, so AHL change is related to the copy number of both plasmid1 and plamid2. However, figure13 shows that AHL concentration is strong relevant to plasmid1 copy number but week relevant to plasmid2 copy number. The maximal value of AHL concentration is about 7e-5 nm, 3 fold lower than AHL limit, which satisfies our expectations of initial states. The condition of NicA2 is contradicted with AHL. More plasmid2 leads to higher NicA2 quantity and the maximal concentration can reach 1.2 nM.
Adding nicotine
We select the initial state after 200000 s as the initial condition for simulation with nicotine. The initial concentration of extracellular nicotine is set to 0.00616 M and the simulation runs for 200000s to get complete dataset of all species change.

Figure 15 GFP with plasmid number

Figure 16 AHL with plasmid number

Figure 17 NicA2 with plasmid number

Figure 18 Product with plasmid number
We mainly analyze the change of GFP/AHL/NicA2/3-Succinoyl-Pyridine with different combinations of plamid1/plasmid2 copy number. The tendency of GFP and AHL change is very similar. Both species are sensitive to the increase of plamid1 copy number. To acquire AHL over 0.1 nm, the copy number of plasmid1 is limited to be smaller than 50 per cell. In consideration of GFP expression level, the requirements of plasmid1 copy number is to be smaller than 50 per cell as well. Figure3 and figure4 show that plasmid1 copy number over 40 per cell leads to low quantity of NicA2 and 3-Succinoyl-Pyridine. If plasmid1 copy number is smaller than 40 per cell, the NicA2 concentration rises with the increase of plamid2 copy number in a linear way. But for 3-Succinoyl-Pyridine, the decline of plasmid1 copy number and increase of plasmid2 copy number together causes more nicotine transformed to 3-Succinoyl-Pyridine.
Conclusions about plasmid copy number
Under the determined combinations of promoters, it is better to choose the plasmid whose copy number is lower than 20 per cell as the backbone of plasimd1. As for plasmid2, copy number ranging from 20 to 40 per cell is an economical option for exact expression control and efficient nicotine degradation.
Optimized conditions for experiment design
Defined parameters:
Promoter 1 strength 0.5
Nicotine input 0.00616 M
Variable parameters:
| Plasmid copy number | Minimal value (per cell) | Maximal value (per cell) | Example |
|---|---|---|---|
| Plasmid 1 | 1 | 20 | pET28 |
| Plasmid 2 | 20 | 40 | pET22b(+) |
| Promoter strength | Minimal value | Maximal value | Example |
|---|---|---|---|
| Promoter 2 (hdnoR) | 0.1 | 0.3 | J23117 |
| Promoter 3 (LuxR) | 0.1 | 1 | J23116 |
