|
|
| (61 intermediate revisions by 2 users not shown) |
| Line 3: |
Line 3: |
| | <style> | | <style> |
| | | | |
| − | .pictureTitle{
| + | .pictureTitle{ |
| − | background: linear-gradient(rgba(0,0,0,.5), rgba(0,0,0,.8)), url("https://static.igem.org/mediawiki/2018/b/bf/T--Munich--background_results.png");
| + | background: linear-gradient(rgba(0,0,0,.5), rgba(0,0,0,.8)), url("https://static.igem.org/mediawiki/2018/b/bf/T--Munich--background_results.png"); |
| − | background-repeat: no-repeat;
| + | background-repeat: no-repeat; |
| − | background-size: cover;
| + | background-size: cover; |
| − | background-position:center;
| + | background-position:center; |
| − | }
| + | } |
| − |
| + | |
| | | | |
| | + | #accordion .card { |
| | + | margin-bottom: 10px; |
| | + | border: 2px solid rgba(0,0,0,.4); |
| | + | } |
| | + | #accordion .card-header { |
| | + | padding:0; |
| | + | background-color:#00A7D4; |
| | + | } |
| | + | #accordion h5 { |
| | + | padding:0; |
| | + | } |
| | + | #accordion .card button{ |
| | + | width: 100%; |
| | + | text-align: left!important; |
| | + | } |
| | + | #accordion button span { |
| | + | font-size: 1.5rem; |
| | + | } |
| | + | |
| | + | |
| | + | @media only screen and (max-width: 809.99px) { |
| | + | #accordion button span { |
| | + | font-size: 1rem; |
| | + | } |
| | + | } |
| | | | |
| | @media only screen and (max-width: 575.98px) {} | | @media only screen and (max-width: 575.98px) {} |
| Line 26: |
Line 50: |
| | Results | | Results |
| | </div> | | </div> |
| − | <h4>First Blick of Phactory</h4> | + | <h4></h4> |
| | | | |
| | </div> | | </div> |
| | | | |
| − |
| + | |
| | <div class="phaContainer"> | | <div class="phaContainer"> |
| | <aside id="phaContentsOuter"> | | <aside id="phaContentsOuter"> |
| Line 39: |
Line 63: |
| | | | |
| | <main class="post-content"> | | <main class="post-content"> |
| − |
| + | <!--- |
| − | <br> | + | <div class="row"> |
| − |
| + | <div class="col-12 text-center"> |
| − | <div class="row justify-content-center"> | + | <figure class="figure" > |
| − | <div class="col-12"> | + | |
| − | <figure class="figure" > | + | |
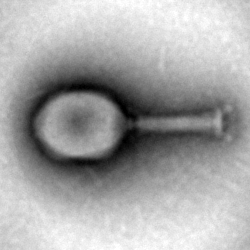
| | <img src="https://static.igem.org/mediawiki/2018/2/2b/T--Munich--MS_3S_phage.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/2/2b/T--Munich--MS_3S_phage.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | <figcaption class="figure-caption">BIG BROTHER IS WATCHING YOU</figcaption> | | <figcaption class="figure-caption">BIG BROTHER IS WATCHING YOU</figcaption> |
| Line 50: |
Line 72: |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | ---> |
| | | | |
| − | <h2> | + | <h2>Optimizing a Cell-Free Expression System</h2> |
| − | Optimizing a Cell-Free System | + | <div class="row"> |
| − | </h2> | + | <div class="col-12"> |
| | <p> | | <p> |
| − | The first step of our project is the optimization of a cell-free expression system as a manufacturing platform for bacteriophages. For our purpose, it was necessary to produce a high-quality cell extrac, in a reproducible and easy manner. Considering a possible commercial use of our product we deemed it necessary to test the potential of upscaling of our preparation protocol.</p> | + | The first part of our project is the optimization of a cell-free expression system as a manufacturing platform for bacteriophages. For our purpose, it is necessary to produce a high quality cell extract, in a reproducible and easy manner. |
| − | <p> | + | <br> |
| − | We decided on the following optimization goals: | + | We focused on achieving the following goals for our cell extract: |
| | </p> | | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| Line 64: |
Line 90: |
| | <li>increase protein content</li> | | <li>increase protein content</li> |
| | <li>find reproducible methods of quality control</li> | | <li>find reproducible methods of quality control</li> |
| − | <li>test cell cultivation in a bioreactor to enable upscaling</li>
| |
| − | <li>find optimal lysis conditions, that produce high-quality extract</li>
| |
| − | <li>test upscaling of cell lysis</li>
| |
| | <li>produce cell-extract that allows phage assembly</li> | | <li>produce cell-extract that allows phage assembly</li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | <br> |
| | + | Considering a possible commercial use of our product we decided to furthermore evaluate the potential for scaling up of our preparation protocol. |
| | + | <br> |
| | + | We chose to try several different approaches to achieve these goals |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>test cell cultivation in a bioreactor to enable upscaling</li> |
| | + | <li>find optimal lysis conditions that produce high-quality extract</li> |
| | + | <li>test scaling up of cell lysis</li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | <br> |
| | + | We found that: |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>bioreactor cultivation allows for upscaling of cell extract production</li> |
| | + | <li>sonication and lysozyme improve the performance of cell extract</li> |
| | + | <li>cultivation and extract preparation barely impacts cell extract composition </li> |
| | + | <li>lyophilization is a good choice for cell-extract storage</li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | + | <!--- |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>biorector cultivation, sonication, lysozyme enhance the performance</li> |
| | + | <li>different cultivation and cell disruption methods do not overly impact the protein composition, but probably the activity of the expressed protein</li> |
| | + | <li>lyophylization is a good way to store cell extract</li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | + | ---> |
| | + | |
| | | | |
| | <hr> | | <hr> |
| | + | |
| | <h3> | | <h3> |
| − | Cultivation | + | Bioreactor Cultivation Allows For Upscaling Of Cell Extract Production |
| | </h3> | | </h3> |
| − |
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Cultivation is the first step in cell extract preparation. The original cell extract preparation protocol uses shaking flask cultivation for biomass production and states that cell harvest at OD 1,8-2,0 is strictly required to produce high-quality extract. To be able to transfer this to the bioreactor we first obtained growth data for both shaking flask and bioreactor cultivation. | + | <p>Cultivating bacteria is the first step in cell extract preparation. The cell extract preparation protocol of Sun et al. uses shaking flask cultivation for biomass production. It recommends cell harvest in the mid-log growth phase at OD 1.8-2.0 to produce high-quality cell extract. We recorded growth curves for shaking flask cultivation and bioreactor cultivation in a lab-scale bioreactor, to compare biomass production under different culture conditions. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 86: |
Line 162: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/2/2b/T--Munich--WL2_fermenter.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/2/2b/T--Munich--WL2_fermenter.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">Growth curves for cell cultivation in a shaking flask and a 2 L bioreactor. Growth curves were approximated by fitting a logistic function (line).</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| − | <hr>
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>The growth curve from shaking flask cultivation showed us that harvest at OD 1,8-2 correlates to the mid-to late logarithmic growth phase. In bioreactor fermentation this correlates to OD between 4 and 6. We decided to test which of these ODs is best to harvest cells for cell extract preparation. | + | <p>The growth curve from shaking flask cultivation, revealed that the mid-log growth phase correlates to an OD of around 2. In bioreactor cultivation the mid-log growth phase is prolonged, due to the better aeration of the medium, and corresponds to an OD between 4 and 6. We harvested cells at OD 4, 5 and 6 to test which harvest point is most suitable to produce high quality cell extract. |
| | </p> | | </p> |
| | | | |
| Line 102: |
Line 176: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/0/02/T--Munich--WL2_fermenter2.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/0/02/T--Munich--WL2_fermenter2.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">FI shaking flask vs fermenter</figcaption> | + | <figcaption class="figure-caption">Protein content (grey) and fluorescence intensity (blue) in cell extract in different cultivation conditions.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | The test reveals that the protein content is higher when cells where harvested at higher OD, with OD 5 yielding the highest protein content. In addition, the cell extract from OD 5 produces the highest fluorescence intensity in the TXTL test. Compared to a cell extract from shaking flask cultivation the signal is reduced to about 60 %. The most likely cause for this result, is that a higher fraction of proteins in the OD 5 cell extract is inactive, possibly due to protein damage during cell lysis (at the time of this experiment, cell lysis was not yet fully optimized). |
| | + | </p> |
| | + | <p> |
| | + | The comparison cultivation methods revealed that cultivation in a bioreactor is an applicable alternative to shaking flask cultivation and offers great potential for upscaling of cell extract preparation. In our 2 L lab-scale bioreactor we were able to produce 20 g of cell pellet, compared to 4-5 g that 2 L of shaking flask cultivation yield. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| | + | <hr> |
| | | | |
| | + | <h3> |
| | + | Sonication and Lysozyme Improve the Performance of Cell-Extract |
| | + | </h3> |
| | + | <h5>Sonication</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
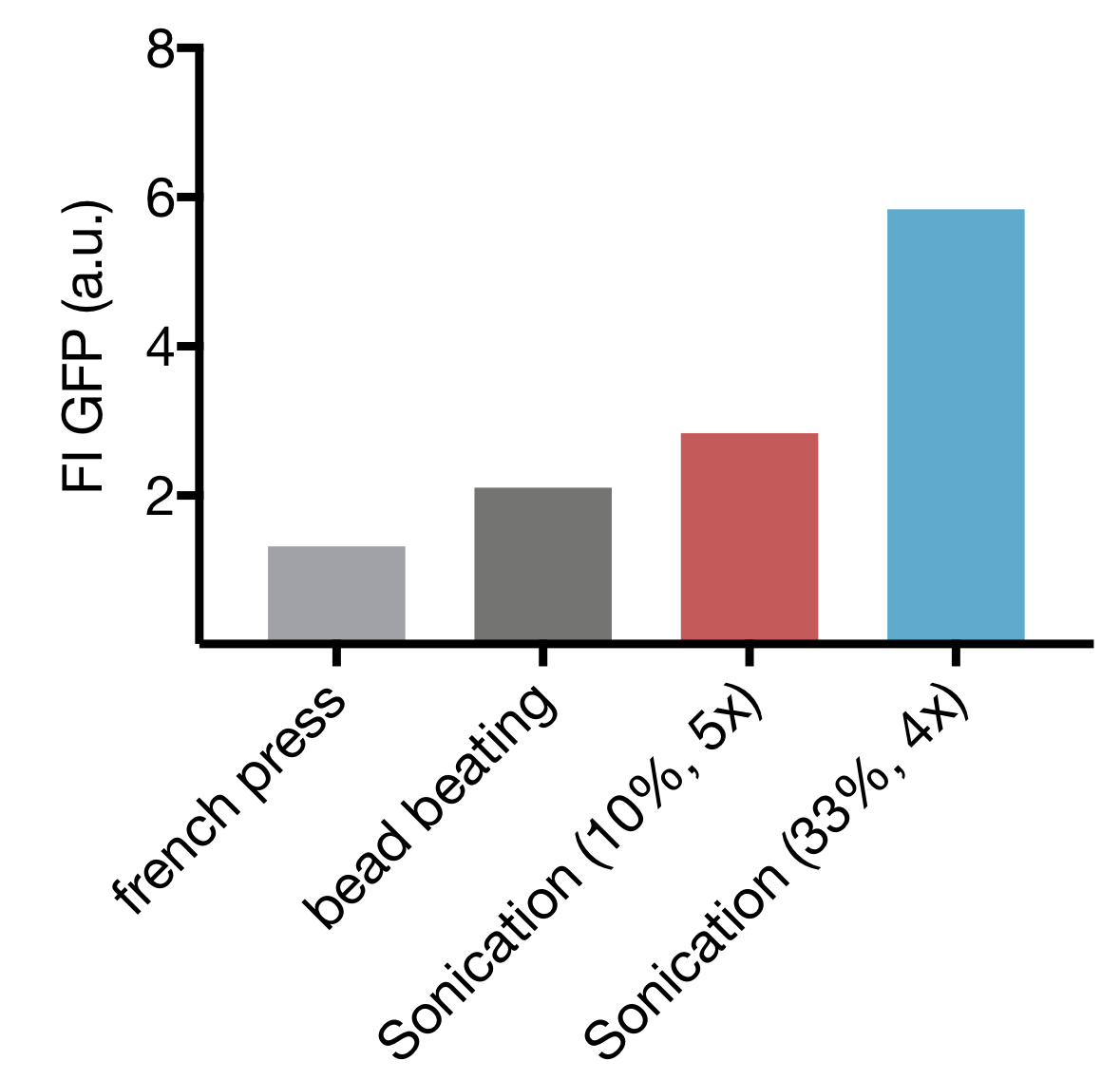
| | + | <p>We tested three different cell lysis methods: beat beating, French press and sonication. Beat beating is an inexpensive and widely employed method but upscaling is problematic and the preparation is tedious and time-consuming. High-pressure cell disruption in a so called ‘French-press’ is also used but these devices are very expensive and not widely prevalent. This contradicts our effort to provide a generally-applicable protocol for preparation of home-made cell extract. A sonication device on the other hand is available in most biochemical labs as it is a commonly used method for cell lysis in protein purification protocols. Nevertheless, we started by comparing all three lysis methods with settings commonly applied for cell lysis. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>Our initial test showed that cell lysis by sonication yields extract with up to 2.8 and 4.4 fold higher expression than cell lysis by bead beating and French press respectively. These findings, combined with the limited options of optimization for both bead beating and French press lysis as well as restricted potential for scaling up of bead beating lead to the decision to focus our optimization efforts on sonication as lysis method.</p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/f/fc/T--Munich--Lysis_methods.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"><p>Comparison of different cell lysis methods for cell extract preparation.</p></figcaption> |
| | + | </figure> |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p>Cell lysis by sonication has two main parameters that can be varied: the amplitude, which correlates to the power emitted by the sonication device and cycle number, representing the total time of sonication. Based on Yong-Chan Kwon & Michael C. Jewett we decided to use 10 s pulses with 15 s pause between pulses to prevent excessive heating of the sample. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/f/f6/T--Munich--Results-WL2_NEW_Protein_Content.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/9/97/T--Munich--Results-WL2_NEW_mTurq.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <figcaption class="figure-caption">Screening of different amplitudes and cycle numbers for sonication.</figcaption> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | <br> |
| | + | Our screening revealed that increase of amplitude as well as increase of cycle number leads to higher protein content. The TXTL test showed high fluorescence intensity only for settings with low cycle numbers. This indicates that prolonged sonication damages the cellular machinery, declining the ability for protein expression. |
| | + | </p> |
| | <p> | | <p> |
| − | This experiment gave 2 important results:
| + | The highest-performing extracts where those obtained by sonication at 4 cycles at 33 % amplitude and 5 cycles at 10 % amplitude. Therefore we focused our next steps on further optimization of those settings. |
| | </p> | | </p> |
| | | | |
| | + | <h5>Lysozyme</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p>In the last step we made an effort to scale up the sonication step. Preliminary experiments (results not shown) indicated 4 mL as the maximal sample volume that could efficiently be lysed with the device we had at our disposal. As indicated in the publication by Yong-Chan Kwon & Michael C. Jewett2 the number of sonication cycles was increased in proportion to the increase of sample volume. The presented results shown in the figure below shows that this approach was successful: Using higher sample volumes for sonication increased protein content and expression quality of the cell extract. This proves that upscaling of cell extract preparation with cell lysis by sonication is indeed feasible. |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/52/T--Munich--Results_Wl2_lysoprot.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/a/ae/T--Munich--Results_Wl2_lysofluo.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <figcaption class="figure-caption">Comparison of cell lysis conditions: increase of sample volume and use of lysozyme.</figcaption> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | <br> |
| | + | Even though previous attempts by Shresta et al.[ref 3] to use Lysozyme in cell extract preparation have been unsuccessful, we tested the effect of adding the cell-wall degrading enzyme at 1 mM concentration to the cell lysis reaction. In our experiments lysozyme increases the efficiency of cell lysis for all tested settings. Moreover, the expression quality was likewise increased after Lysozyme addition, indicating that the remaining enzyme does not interfere with protein biosynthesis. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <h5>Cell Extract Processing</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | Cultivation and cell lysis are the apparent most crucial steps in preparation of cell extract but there are other steps that can have a great impact onto cell extract quality. Some of our findings regarding those steps are: |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>Freezing cell pellets overnight for next-day processing does not interfere with extract quality but confers a major simplification of cell extract preparation</li> |
| | + | <li>Preliminary results indicate that omitting the run-off reaction after lysis does not significantly impair the extract quality</li> |
| | + | <li>Dialysis of cell extract can be substituted by diafiltration in centrifugal filters without reducing expression yield of the resulting extract </li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <hr> |
| | + | |
| | + | <h3>Characterizing Cell Extract Quality </h3> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | <ul style="">
| + | In order to validate the success of our optimization efforts, adequate characterization of cell extract is indispensable. |
| − | <li>the optimal OD to harvest culture from the bioreactor is around 5. This gives the highest protein content and also the best expression.</li>
| + | Here we will give a short review about the tools for quality assessment we implemented. Further details about characterization |
| − | <li>preparing cell extract from bioreactor cultivated cells under comparable conditions gives equal quality extract than cell extract from shaking flask cultivated cells.</li>
| + | of these tools are available on the <a href="https://2018.igem.org/Team:Munich/Measurement">Measurement</a> page. |
| − | </ul> | + | |
| | </p> | | </p> |
| | + | <p> |
| | + | We had initially defined increase of protein content in cell extract as optimization goal. However we realized early on, that protein content alone is no measure for extract quality, because it provides no information about the activity of the cellular machinery. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | <p> | | <p> |
| − | In our small lab-scale bioreactor with 2 L cultivation volume we were able to obtain 20 g cell pellet. 2 L cultivation in shaking flasks only yields 4,5 g pellet.
| + | To assess functionality of cell extract, testing of protein expression is necessary, which can be achieved by expressing fluorescent proteins and measuring the fluorescence time trace in a plate reader. To find the optimal quality control we compared the performance of different fluorescent proteins in cell extract. |
| | </p> | | </p> |
| − | <hr> | + | <p> |
| − | <h3> | + | We chose expression of mTurquoise – a variant of Cyan Fluorescent Protein (CFP) – as our favorite quality control for cell extract, as it results in high fluorescence intensity in a reproducible manner. |
| − | Lysis
| + | </p> |
| − | </h3> | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/e/e0/T--Munich--Results-WL2_Four_Fluorescence.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p> |
| | + | In addition, to examining protein expression we decided to analyze transcription uncoupled from translation by transcribing an RNA aptamer that binds a fluorescent dye. |
| | <p> | | <p> |
| − | Of the three commonly used methods of cell lysis, each has teir advantages and drawbacks. For cell extract preparation bead beating is most established, but the caveat there, is that it doesn’t leave room for upscaling, besides being tedious and time-consuming. Therefore, we decided early on to focus our optimization approach on Sonication as method for cell lysis.
| + | We compared the fluorescence time trace of a Malachite Green binding aptamer in different cell extract samples. The fluorescence time traces decline after 30-50min indicating, that RNA degradation starts to prevail over transcription. Differences in the observed kinetics can be explained by variations in cell extract composition. |
| | </p> | | </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/e/e3/T--Munich--WL2_MG_aptamer.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | <div class="row">
| + | <h3> |
| | + | Cultivation and Extract Preparation Barely Impacts Cell Extract Composition |
| | + | </h3> |
| | + | |
| | + | |
| | + | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p> |
| | + | The main question was, if the different performance of the cell extracts (P15, E10, myTXTL), especially concerning phage titers, is due to a divergent composition or caused by a discrepancy in activity of the relevant proteins e.g. transcription and translation related proteins |
| | + | Therefore, one student was sent to Bavarian Biomolecular Mass Spectrometry Centre (BayBioMS), where he analyzed the samples under the supervision of Dr. Christina Ludwig. |
| | + | </p> |
| | + | <p>The results of the Mass Spectrometry gave us insight into the composition of the different TX-TLs. We were able to identify 1771 proteins in all the extracts. The results indicate a high homogeneity between all three TX-TLs, as illustrated in the heatmap. |
| | + | Based on the LFQ values (Label Free Quantification) a volcano plot of two samples (Arbor/E10, Arbor/P15 and E10/P15) was generated. In general, there is no protein more frequent in Arbor TX-TL in comparison to E10 and P15. In contrast, some translation related proteins were slightly more abundant in E10 and P15 like certain tRNA ligases (PheS and PheT). The RecBCD subunits (the dsDNA degrading complex) is of similar abundance in all TX-TLs, showing the importance to consider its negative impact on the assembly. The results were further validated with DAVID, a tool for finding metabolic pathways based on proteomic data. The identified pathways indicate no correlation with transcription/translation related pathways (data not shown). |
| | + | </p> |
| | + | <p> |
| | + | We concluded that the performance difference is most likely due to decreased activity during TX-TL preparation rather than a change in composition. |
| | </p> | | </p> |
| − |
| |
| | </div> | | </div> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <figure class="figure">
| + | <figure class="figure"> |
| − | <img src="https://static.igem.org/mediawiki/2018/a/ae/T--Munich--Results_Wl2_lysofluo.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | + | <img src="https://static.igem.org/mediawiki/2018/5/5f/T--Munich--Results_Wl3_heatmap.png " class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption">Illustration of the abundance of all sequenced proteome of duplicates from P15, E10 and myTXTL based on LFQ values. Heatmap of the label-free quantification intensities of all proteins identified. Diagram shows duplicate samples from the three cell extracts analyzed. Green indicates high expression, red indicates low expression. </figcaption> |
| | + | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| − | <hr>
| + | |
| | + | |
| | <div class="row"> | | <div class="row"> |
| − | <div class="col-12 col-md-6"> | + | <div class="col-12 col-md-4"> |
| − | <p>Discussion goes here | + | <figure class="figure"> |
| − | </p> | + | <img src="https://static.igem.org/mediawiki/2018/a/a2/T--Munich--Results_arbor_p15_volcano.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Arbor p15</figcaption> |
| | + | </figure> |
| | </div> | | </div> |
| − | <div class="col-12 col-md-6"> | + | <div class="col-12 col-md-4"> |
| | <figure class="figure"> | | <figure class="figure"> |
| − | <img src="https://static.igem.org/mediawiki/2018/5/52/T--Munich--Results_Wl2_lysoprot.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | + | <img src="https://static.igem.org/mediawiki/2018/d/d8/T--Munich--Results_arbor_e10_volcano.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption">Arbor e10</figcaption> |
| | + | </figure> |
| | </div> | | </div> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/0/0c/T--Munich--Results_e10_p15_volcano.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">e10 p15</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12"> |
| | + | <figcaption class="figure-caption">Volcano plots of myTXTL vs. P15 (A), myTX-TL vs. E10 (B) and E10 vs. P15 (C), LC-MS/MS measurements of the E. coli proteome lysates were performed in duplicates on a Q-Exactive HFX instrument in data-dependent acquisition mode (Top18 method). Total protein injection amount was 0.5 µg. Gradient length was 1h. The data was then analyzed using MaxQuant and the Uniprot E. coli K12 fasta file. The LFQ data was filtered for proteins that occur in both duplicates of at least one cell lysate group (Arbor E10 P14).</figcaption> |
| | + | </div> |
| | </div> | | </div> |
| | | | |
| | <hr> | | <hr> |
| | + | |
| | <h3> | | <h3> |
| − | Processing
| + | Lyophilization Is a Good Choice for Cell-Extract Storage |
| | </h3> | | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | The suggested steps in cell extract processing are:
| + | With regard to global application of Phactory it is crucial to enable long-term storage and shipping of our cell extract. In order to fulfill these requirements we chose to freeze-dry our cell extract. Lyophilized extract can be stored at ambient temperature and can be reactivated by addition of only nuclease-free water. |
| | + | |
| | + | Initial tests for the optimal lyophilization conditions revealed several important points: |
| | </p> | | </p> |
| − | <p>
| |
| | <ul style=""> | | <ul style=""> |
| − | <li>run-off reaction</li> | + | <li>cell extract quality is only preserved when cell-extract is mixed with the TXTL reaction buffer prior to lyophilization</li> |
| − | <li>dialysis</li> | + | <li>the preservation of quality does not depend on the size of lyophilized extract aliquots</li> |
| − | <li>storage at -80°C</li>
| + | |
| | </ul> | | </ul> |
| | + | <br> |
| | + | <br> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>We tested the expression quality of several of our home-made cell extracts from fresh and lyophilized aliquots. This comparison also includes the commercially available myTXTL® cell-free expression system (arbor biosciences) and highlights two important results of our efforts to optimize production of cell extract: |
| | </p> | | </p> |
| | + | <ul style=""> |
| | + | <li>our improved cell extract can be lyophilized and retains 40-70 % of its performance</li> |
| | + | <li>our final settings for cell extract preparation yield extract that clearly outperforms the commercial system, with up to 9-fold higher protein expression</li> |
| | + | </ul> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/4/49/T--Munich--Results-WL2_LYO_vs_FRESH.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Our two tested samples of cell extract retained 70 and 90 % of expression quality respectively after lyophilization.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| | + | <h2> |
| | + | Synthetic Phage Manufacturing |
| | + | </h2> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | We found that the run-off reaction as suggested in the <a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3960857/">Sun et.al paper</a> is indeed beneficial and that dialysis is superior to diafiltration via centrifugal filters.
| + | Phages were manufactured based on three components: the cell extract, an energy solution (ATP, GTP, NAD+ etc.) and a supplement |
| | + | solution (aminoacids, tRNAs, pholic acid etc.). The only additional component for phage assembly is pure phage DNA. We expanded the assembly platform by |
| | </p> | | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>producing the first clinically relevant phages at therapeutic concentrations</li> |
| | + | <li>manufacturing of therapeutic phages independent of a living pathogen</li> |
| | + | <li>achieving similar phage titers as the commercial cell extract</li> |
| | + | <li>determining the amount of DNA necessary for sufficient phage assembly</li> |
| | + | <li>calculating the amount of DNA produced in the cell extract by replication</li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | <h3> | | <h3> |
| − | Quality Control
| + | DNA Purification |
| | </h3> | | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p> An essential part of the bacteriophage manufacturing is the DNA. For the extraction of the bacteriophage DNA we started with a Phenol-Chloroform extraction protocol (link). To purity was controlled with an agarose gel, where two distinct bands where present in for all four samples the proper high mass band and a low mass band. |
| | + | </p> |
| | + | <p> To reduce the amount of the low mass bands the protocol was changed to a column based technique (link). With this technique the incorrect band where significantly reduced in all samples, which was proven with an agarose gel.</p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/f/fe/T--Munich--agarose_gel_dirty_phage.jpeg " class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> The agarose gel shows the DNA of a phenol chloroform purification of the bacteriophage genomes from left to right T7, GEC-3S, T4 and NES. The ladder in the first lane from left is the 1kb extended range ladder from NEB.</figcaption> |
| | + | </figure> |
| | + | |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/77/T--Munich--agarose_gel_clean_phage.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> The agarose gel shows the DNA of bacteriophages which was column purified. The bands are from left to right, a the 1kb extended range ladder from NEB the CLB-P2, CLP-P3, T4, T5, T7, NES, FFP and GEC-3S bacteriophage. |
| | + | </figcaption> |
| | + | </figure> |
| | + | |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | | | |
| | <h3> | | <h3> |
| − | Storage
| + | Home-Made Cell Extract Achieves Phage Titer Comparable to the Commercial System |
| | </h3> | | </h3> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>We determined the concentration of phages assembled in our cell extracts P15 and E10, which had the most promising |
| | + | results in our cell extract quality control. We compared our extracts to the commercial extract (myTXTL) and saw a similar performance. |
| | + | </p> |
| | <p> | | <p> |
| − | For long term storage of our cell-extract we decided to try lyophilization. After initial tests we found that:
| + | All in all our extract has the same phage assembly potential as the commercial cell extract. Moreover, lyophylization, which makes |
| | + | the cell extract more long lived, does not reduce the effiency of bacteriophage assembly. We concluded that phages could be assembled on site in a durable lyophilized cell extract. |
| | </p> | | </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/79/T--Munich--Results_Phage_Titer.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Comparison of assembled T7 phage in our extracts (P15 and E10) in comparison to the commercial extract (myTXTL). Blue bars indicate an assembly within fresh extract, whereas red indicates an assembly in a lyophilized cell extract.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| | | | |
| | + | <hr> |
| | | | |
| | + | <h3> |
| | + | Bacteriophages Can Be Assembled Independent of the Living Host |
| | + | </h3> |
| | | | |
| − | | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | <ul style="">
| + | Phactory has is the ability to assemble various bacteriophages, in a bacteria-independent manner. To underline this feature and demonstrate universal applicability, we assembled a variety of different E. coli phages, both DNA and RNA-based. |
| − | <li>cell extract quality is only preserved when cell-extract is mixed with the txtl- reaction buffer prior to lyophilization. </li>
| + | |
| − | <li>the retention of quality does not depend on the size of lyophilized extract aliquots.</li>
| + | |
| − | <li>We then tested the expression quality of several of our home-made cell extracts from fresh vs. lyophilized aliquots.</li>
| + | |
| − | </ul>
| + | |
| − | </p>
| + | |
| − | <p>
| + | |
| − | plot: lyophilisation
| + | |
| | </p> | | </p> |
| | + | <br> |
| | + | <br> |
| | + | </div> |
| | + | </div> |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Cultivation is the first step in cell extract preparation. The original cell extract preparation protocol uses shaking flask cultivation for biomass production and states that cell harvest at OD 1,8-2,0 is strictly required to produce high-quality extract. To be able to transfer this to the bioreactor we first obtained growth data for both shaking flask and bioreactor cultivation. | + | <p>The successful assembly of all phages was confirmed by plaque assay and transmission electron microscopy (TEM). In addition, DNA encoding for NES and FFP phages was used to perform assembly of these phages in our cell extract. However, we were not in possession of the respective host bacterial strains and therefore could not demonstrate successful assembly. |
| | </p> | | </p> |
| | + | |
| | + | <p> |
| | + | Pathogenic bacteria such as salmonella, pseudomonas and staphylococcus are prone to develop multi-drug resistance and pose an urgent or serious threat (Centers for Disease Control and Prevention, 2013. Antibiotic/Antimicrobial Resistance.). Therefore, to fulfill this medical need, phages specific for these bacteria should be assembled next in our cell-free system. Potentially, this would require co-expression of the respective sigma factors that are needed for initiation of transcription. |
| | + | </p> |
| | + | |
| | </div> | | </div> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| | <figure class="figure"> | | <figure class="figure"> |
| − | <img src="https://static.igem.org/mediawiki/2018/8/8a/T--Munich--Results_lyophilization.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | + | <img src="https://static.igem.org/mediawiki/2018/7/76/T--Munich--Results--WL3_Titer_difference.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption>
| + | <figcaption class="figure-caption"> Overview of the successfully assembled phages. MS2 (RNA-phage), T4 (DNA phage), T5 (DNA phage), T7 (DNA phage), CLB-P2 (clinically relevant), CLB-P2 (clinically relevant), GEC-3S P2 (clinically relevant).</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 218: |
Line 557: |
| | | | |
| | | | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/2/29/T--Munich--Results-classaverage_MS2.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage MS2.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/3/32/T--Munich--Results-classaverage_T4.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage T4.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/8/8a/T--Munich--Results-classaverage_T5.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage T5.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/a/a4/T--Munich--Results-classaverage_T7.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage T7.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/e/e1/T--Munich--Results-classaverage_P2.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage CLB-P2.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/6/66/T--Munich--Results-classaverage_P3.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage CLB-P3.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/2/2e/T--Munich--Results-classaverage_3S.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Phage GEC-3S.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <figcaption class="figure-caption">TEM class averages of phages assembled in cell extract. We assembled the phages MS2, T4, T5, T7 and the clinically relevant phages CLB-P2, CLB-P3, and GEC-3S.</figcaption> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | <hr> |
| | + | |
| | + | <h3>DNA Concentration Determines Phage Titer</h3> |
| | + | <h5>Bacteriophage Titers Correlate With DNA Concentration</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p>To prove the influence of the DNA concentration on the bacteriophage titer, cell extract reactions were prepared with varying T4 DNA concentrations. The bacteriophage production was performed. The titer of the bacteriophages was measured with the top agar method and the formed plaques were counted. The increase in DNA concentration results also in an increase in the bacteriophage concentration. This increase is nonlinear and our model predicted. This finding is probably due to a critical concentration of phage proteins, which have to be reached for capsid head assembly (similar to a critical micelle concentration). |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/70/T--Munich--Results_DNA_CONC_T4.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Assembly of the T4 phage depending on the DNA concentration.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/0/0d/T--Munich--Results_DNA_CONC_T7.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Assembly of the T7 phage depending on the DNA concentration.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | |
| | + | <h5>Cell-Free Systems Replicate Phage Genomes</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>The DNA sequence added to the cell-free system serves as the template for the required phage. We saw, that DNA-Polymerases can amplify the DNA segment, multiplying the amount of DNA in the cell-free reaction. |
| | + | </p> |
| | + | <p> |
| | + | To assess this effect and its dependence on deoxynucleotide triphosphates (dNTPs), we performed an absolute quantification of T7 DNA in the cell-free reaction by quantitative PCR (qPCR). A standard curve with a serial dilution of T7 DNA. We used the TXTL qPCR protocol (add link). |
| | + | We used two selected cell extracts from us (P15 and E10), which reached similar phage titers compared to the commercial cell extract (myTXTL) in previous experiments. |
| | + | We compared the replication potential in comparison to myTXTL. |
| | + | </p> |
| | + | <p> |
| | + | The addition of dNTP to the reference reaction leads to an increase in DNA concentration by a factor of 15 in the reaction after 4 hours (290 ng compared to 19 ng). This is higher than in the myTXTL reaction without additional dNTPs, in which there is a 1.8-fold increase in DNA (91 ng compared to 51 ng) after the 4-hour reaction. |
| | + | The home-made cell extracts P10 and E15 however do not resemble this behavior. |
| | + | </p> |
| | <p> | | <p> |
| − | Our two tested samples of cell extract retained 70 and 90 % of expression quality respectively after lyophilization.
| + | It would be desirable to increase DNA amplification in our cell extracts. We therefore conducted a cause analysis, focusing on the T7 replication system. A more than 250-fold increase in processivity of the T7 DNA polymerase is achieved by its binding behavior to E. coli thioredoxin1. We suspected reduced presence of this factor in our cell extract. Thioredoxin could be added to a phage assembly reaction to further test these assumptions. However, our proteome analysis did not confirm that there were low levels of thioredoxin present in our cell extract. |
| | </p> | | </p> |
| | | | |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/3/36/T--Munich--Results_WL2_Qpcr.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">DNA replication within cell extracts, E15 and P15 are self prepared cell extracts, which reached similar phage titers compared to the commercial cell extract (myTXTL). ctrl = control without dNTPs. </figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <h3>Modelling Phage Production in Cell Extract</h3> |
| | | | |
| | <h2> | | <h2> |
| − | Bacteriophage Assembly | + | Modular Bacteriophage Composition |
| | </h2> | | </h2> |
| | + | |
| | <h3> | | <h3> |
| − | Home-Made Systems Can Be Compared To Commercial Systems
| + | Protein engineering |
| | </h3> | | </h3> |
| | + | |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | In the TX-TL system should be possible to modify phage proteins without altering their genome. This was attempted by modifying HOC (highly immunogenic capsid protein), which is part of the capsid protein structure of the T4-phage. Therefore, His-TEV-YFP-HOC was separately expressed and the purified protein was applied to our phage assembly. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | For protein modification of the T4 capsid protein HOC, a plasmid for protein expression was cloned and His-YFP-HOC (82kDa) was expressed. The plasmid was transformed into BL21, expressed and puryfied by nickel affinity chromatography and gelfiltration. |
| | + | </p> |
| | + | <p>FILM</p> |
| | + | <p>MODEL</p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row mt-3"> |
| | + | <div class="col-12"> |
| | + | <div id="accordion"> |
| | + | <div class="card" style="background-color:#017FD4;"> |
| | + | <div class="card-header" id="headingOne"> |
| | + | <h5 class="mb-0"> |
| | + | <button class="btn btn-link collapsed" data-toggle="collapse" data-target="#collapseOne" aria-expanded="false" aria-controls="collapseOne"> |
| | + | <span class="pl-3" style="color: white;">Protein Purificaton</span> |
| | + | </button> |
| | + | </h5> |
| | + | </div> |
| | + | <div id="collapseOne" class="collapse" aria-labelledby="headingOne" data-parent="#accordion"> |
| | + | <div class="card-body bg-white"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/7c/T--Munich--Results_purifyHoc.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Elution peaks of His-HOC-YFP from nickel chromatography (A) and gelfiltration (B) with corresponding 15% silverstained SDS-PAGE.</figcaption> |
| | + | </figure> |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | After the two chromatography steps, the purified protein had a final concentration of 70µM and no by-products were visible by silver staining. The CD-spectra (minima of 218nm) of the protein corresponds to the model and the Ramachandran plot indicates that the protein was not denatured during purification. Then, the purified protein was intentionally denatured by thermal transition. Thereby, it was confirmed that the protein is stable below 40°C. This is of importance, as phage assembly is performed at 29°C.</p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row mt-3"> |
| | + | <div class="col-12"> |
| | + | <div id="accordion"> |
| | + | <div class="card" style="background-color:#017FD4;"> |
| | + | <div class="card-header" id="headingTwo"> |
| | + | <h5 class="mb-0"> |
| | + | <button class="btn btn-link collapsed" data-toggle="collapse" data-target="#collapseTwo" aria-expanded="false" aria-controls="collapseTwo"> |
| | + | <span class="pl-3" style="color: white;">Protein Characterization</span> |
| | + | </button> |
| | + | </h5> |
| | + | </div> |
| | + | <div id="collapseTwo" class="collapse" aria-labelledby="headingTwo" data-parent="#accordion"> |
| | + | <div class="card-body bg-white"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/1/10/T--Munich--Results_CD_His_YFP_HOC.PNG" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">CD spectra of His-YFP-HOC with the specific minimum at 218 nm.</figcaption> |
| | + | </figure> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/5f/T--Munich--Results_thermo%C3%BCbergang_His_YFP_HOC.PNG" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Thermal transition of His-YFP-HOC in correlation with the absorbance.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="card-body bg-white"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/9/96/T--Munich--Results_Ramachandran.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Ramachandran plot based on the amino acid sequence.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>The purified protein was added to the assembly mix. Bacteria were transfected with the modified phages. Unbound phages and protein were removed by centrifugation. Fluorescence was measured in dependence of the proximity to the bacteria. Theoretically, YFP intensity should correlate with the binding of YFP-HOC modified phages to the bacteria. </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/f/f2/T--Munich--Results_wl3_sampleT4_XY122-01.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"></figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/b/be/T--Munich--Results_Engineering_negativecontrol-2.png " class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"></figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figcaption class="figure-caption">Bacteria infected with (A) and without (B) YFP modified phages </figcaption> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | The purified protein was added to the assembly mix. Bacteria were transfected with the modified phages. Unbound phages and protein were removed by centrifugation. Fluorescence was measured in dependence of the proximity to the bacteria. Theoretically, YFP intensity should correlate with the binding of YFP-HOC modified phages to the bacteria. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | <hr> |
| | + | |
| | <h3> | | <h3> |
| − | Cell-Free Systems Allow Host Independent Bacteriophage Assembly
| + | Phage Genome Engineering |
| | </h3> | | </h3> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>An additional advantage of Phactory is the possibility of rebooting bacteriophages from their genomic template, which is especially important for the genetic engineering of bacteriophages. With the self-made cell extract it was possible to manufacture an engineered MS2 RNA phage, where a polyhistidine-tag was added on the phage RNA polymerase. The genome template was generated via simple cloning methods such as gibson assembly and PCR amplification. After purification of the engineered genome, phages were assembled in our self-made P15 cell extract. A plaque assay confirmed the successful assembly of functional phages with a titer of 3 × 10<sup>7</sup> PFU/ml.</p> |
| | + | <p>To test the his-tag modification the Phages were amplified in a bacterial culture flask. After lysis of the bacteria the tagged polymerase was purified from the supernatant by nickel affinity chromatography. An SDS-PAGE proved, that the 62 kDa Protein remained in the Nickel column due to the successfully engineering of an inserted His-tag. </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/52/T--Munich--MS2_gel_enge.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure." height=”50%” width="50%" > |
| | + | <figcaption class="figure-caption">SDS gel of the His-tag purified MS2 RNA dependent RNA polymerase</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | |
| | + | <h2> |
| | + | Quality Control |
| | + | </h2> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | Quality control covers several aspects of phage manufacturing including phage functionality, endoxin levels and DNA purity. We found that |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <ul style=""> |
| | + | <li>we are able to produce functional phages in our cell extract</li> |
| | + | <li>the cell extract of our optimized strain has an endotoxin content below the detection limit and our regular self-produced cell extract has fewer endotoxins than the commercial cell extract</li> |
| | + | <li>next-generation sequencing allowed us to accurately quantify contamination</li> |
| | + | <li>phenol-chloroform extraction leads to a large amount of contaminating DNA which complicates phage assembly</li> |
| | + | <li>next-generation sequencing helped us to improve our purification protocols, leading to improved phage assembly</li> |
| | + | </ul> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | <h3> | | <h3> |
| − | Quality Control Of Bacteriophages
| + | Assessing Phage Functionality by Plaque Assays |
| | </h3> | | </h3> |
| − | <p>Discussion goes here | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p>We performed a Plaque Assay to determine the activity of the viable phages (titer) in our assembly batch. By creating serial dilutions, we were able to calculate a plaque forming units/milliliter (PFU/ml) value. The <a href="https://static.igem.org/mediawiki/2018/2/23/T--Munich--AgarOverlayPlaqueAssay_.pdf "> plaque assay protocol </a> was used. |
| | </p> | | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | | | |
| | <div class="row"> | | <div class="row"> |
| Line 242: |
Line 860: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/d/d0/T--Munich--Results_Wl3_T7_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/d/d0/T--Munich--Results_Wl3_T7_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption"> </figcaption> |
| | + | |
| | + | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 249: |
Line 869: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/b/b5/T--Munich--Results_Wl3_MS2_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/b/b5/T--Munich--Results_Wl3_MS2_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption"> Plaque Assay of manufactured T7 DNA -bacteriophage (top) and MS2 RNA-bacteriophage (bottom) in our self-produced cell extract P15</figcaption> |
| | + | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| | | | |
| | + | <!--- |
| | + | <h5> |
| | + | Bacteriophages From Phactory Assemble And Are Fully Functional |
| | + | </h5> |
| | | | |
| − | <h3> | + | |
| − | Assembly Of Clinically Relevant Bacteriophages
| + | <div class="row"> |
| − | </h3> | + | <div class="col-12 col-md-6"> |
| − | <h3> | + | <p>Discussion goes here |
| − | Sequ-Into Complements Continuous Engineering Cycles in Phactory
| + | </p> |
| − | </h3> | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/7f/T--Munich--Results_QPCR_Nils.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">qpcr</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <h5> |
| | + | RNA-Detection via RT-qPCR |
| | + | </h5> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | Wetlab3 sequenced several phage genomes after preparation using ….
| + | To rapidly detect functionality of the phages, Reverse Transcription-quantitative PCR (RT-qPCR) was applied to pellets of T7-infected host cells at different timepoints. The RT-qPCR protocol was used (add link). Using the delta-delta-Ct method3, relative expression was determined and normalized to the value at 3 minutes after addition of the phages. Whereas expression of E. coli genes remains stable, expression of three T7 genes is elevated throughout the experiment. The change of expression of all three phage genes reaches a first peak after 12 minutes and a second peak after 18 or 21 minutes. |
| − | After receiving the phage genomes, one of the first steps to do is analyse the received sequences.
| + | |
| − | This has been done using an inhouse software poreSTAT [1].
| + | |
| − | For each sequencing sample, it is analysed how many bases are sequenced, what bp yield has been achieved and how many pore were used.
| + | |
| − | Considering the sequence in which the reads have been acquired, it can nicely be seen how the used chip gets worn out with each sequencing experiment.
| + | |
| − | While for the T7 sequencing almost all pores are usable, the used pores decreases with every sequencing experiment …
| + | |
| − | _*_*
| + | |
| − | 4 missing figures
| + | |
| − | _*_*
| + | |
| − | While the T7 experiment produced most reads and most base-pairs sequenced, the remaining three experiments produced less data.
| + | |
| − | The read and base-pair yield can be seen in Table 1.
| + | |
| | </p> | | </p> |
| | + | <p> |
| | + | The sharp increase of phage gene expression in the first phase of the experiment displays the ability of the phages to successfully infect the bacteria and initiate reproduction. The second and even higher increase of expression is likely attributed to a second wave of infection of already replicated phages. This indicates that the phages are capable of reproducing inside their host bacteria, resulting in multiplication of functional phages. |
| | + | The increase of expression of T7P01 is more pronounced than that of T7P07, which is in turn is stronger than that of T7P29. This circumstance is likely caused by differences in primer efficacy, a value describing the doubling rate in between every PCR cycle. For better reliability of the RT-qPCR quality control, primer efficacy should be assessed by creating a standard curve. |
| | + | </p> |
| | + | <p>All RNA samples were pooled and checked for DNA contamination, which could have interfered with the RT-qPCR. Absence of DNA was determined electrophoresis in a gel electrophoresis</p> |
| | + | <br> |
| | + | <br> |
| | + | </div> |
| | + | </div>---> |
| | | | |
| − | <p>TABLE</p> | + | <h3>Assessing Endotoxin Levels</h3> |
| | + | <h5>Msb-B Knockouts Reduce Endotoxin Levels By 49-Fold</h5> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>Endotoxins are pyrogens deriving from gram-negative bacteria. Their mini from any pharmaceutical product is mandatory. Therefore, or Phactory, we engineered an E. coli strain lacking lipid A, a major endotoxin component and used this bacterium to produce our cell extract To evaluate endotoxin content of different cell extracts, a Limulus Amebocyte Lysate (LAL)-test was performed according to the <a href="https://www.genscript.com/product/documents?cat_no=L00350&catalogtype=Document-PROTOCOL">supplier manual</a>. As a reference, we compared the cell extract from our msbB-deficient strain (K2) to a cell extract from a wild-type strain (K4) as well as a commercial cell-free system (myTXTL, Arbor Biosciences). A solution with live <i>E. coli</i> served as a positive control.</p> |
| | + | <p>Compared to the K4 strain our msbB-deficient K2 cell extract had 49-fold reduced endotoxin levels (0.06 EU/ml compared to 2.94 EU/ml). Other cell extracts such as the P15 cell extract (3.83 EU/ml) and the commercial myTXTL (4.65 EU/ml) had even higher endotoxin contents.</p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/53/T--Munich--Results_Wl2_LAL.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Endotoxin content in different cell-extracts determined by LAL-Test. Error bars indicate standard deviation of the measured plateau values. Error bars indicate SD</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | <p>The actual task here was to assemble the phage genomes from the Nanopore reads. | + | <div class="row"> |
| − | Particularly Nanopore sequencing is well suitable for genome assembly, since its long reads allow to reduce ambiguity of highly similar and repetitive sequences.
| + | <div class="col-12"> |
| − | Figure 2 shows the reads distributions of the sequencing experiments.
| + | <p>A calibration curve using known endotoxin concentrations is required for the LAL-Test. A dilution series ranging from 0.625 EU/ml to 5 EU/ml. The fitting curve is used to interpolate the concentrations in the unknown sample. The linear fit of the calibration curve had a R2 of 0.98, an intersection with the y-axis at 0.38 and a slope of 0.39 ml/EU. These values are in accordance with the requirements of the LAL-Test manufacturer. </p> |
| − | _*_*
| + | </div> |
| − | 4 missing figures
| + | </div> |
| − | _*_*
| + | |
| − | However, during the initial screening it could be seen, that the read lengths do not approach the thought length of the phage genomes in the 50-70kbp range for T7 and 100 kbp range for the remaining phages.
| + | |
| − | Smaller reads generally lead to more ambiguity and thus are to be avoided from a bioinformatics point of view.
| + | |
| − | However, since no bioinformatician can change the sequenced data afterwards, we went on with the given data.
| + | |
| − | There are several tools available for de-novo genome assembly in general, however most approaches are used for short-read genome assembly (2nd generation sequencing, Illumina) and employ a de bruijn graph approach.
| + | |
| − | Here we have 3rd generation sequencing data which must be handled totally different from old short-read sequencing data: the reads are less perfect in terms of sequencing errors. While short-reads nowadays have error-rates of about 1% (e.g. 1 base out of 100 is reported incorrectly), this error is up to 15% for nanopore sequencing data using newest sequencing chemistry (R9.4 at the time of wiki-freeze).
| + | |
| − | Thus the number of available assemblers drops dramatically, where canu [2] and miniasm [3] are the most prevalent ones. Both rely on a overlap-layout-consensus approach, which, historically, can be seen as the father of all assemblers (see celera assembler [5]).
| + | |
| − | We first used canu to assemble our genomes. Unfortunately we have been confronted with a major problem: contamination. We thus developed sequ-into to first detect the contamination and also get rid of contamination-originated reads.
| + | |
| − | More on the performance and finding while using sequ-into can be found at …[link to /Software].
| + | |
| − | Particularly for assembly, contamination is bad because it can lead the assembler into wrong directions – depending on the phylogenetic distance of the original sample and the contamination.
| + | |
| | | | |
| − | After getting rid of the contamination we noticed some strange patterns in the phage genome assemblies after re-aligning the reads to the assembly, which can be seen in Figure 3.
| + | <div class="row"> |
| − | *_*_
| + | <div class="col-12"> |
| − | missing figure
| + | <p>Removal of endotoxins is impeded by their tendency to form stable interactions with other biomolecules2. Our method of preventing the lipid A biosynthesis is therefore superior to extensive isolation steps required for removing endotoxins in conventional phage production. </p> |
| − | *_*_
| + | </div> |
| | + | </div> |
| | + | <h3> |
| | + | Nanopore Sequencing enhances DNA purification |
| | + | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>Besides the cell extract, the DNA quality is the key to phage assembly. Unpure DNA (Host DNA, proteins contamination) inhibits the assembly of the phage within the cell lysate. |
| | </p> | | </p> |
| | | | |
| − | <p> | + | <p>Our first attempts with classical chloroform/phenol extraction failed because of low molecular DNA (agarose gel). Therefore, we decided to search for other purification protocols, DNAseI and the Norgene kit (46800) tremendously increased DNA purity. With this DNA, we were finally able to achieve high titers by assembly. |
| − | In theory we can expect a uniform coverage over the full genome, since there is no bias for read template generation during sample preparation (thanks to random primers).
| + | |
| − | However, what we saw here is that the first part has lower coverage than the remaining part and there is a high-coverage region in the middle of the assembled genome.
| + | |
| − | Since we know phage genomes may have repitions, we thought to splite the genome in the middle and reorganise the structure to no avail.
| + | |
| − | We thus tried to use the other assembler, miniasm, which is known for very fast assemblies, with little error correction.
| + | |
| − | However, this error correction can be achieved by combining miniasm with minimap for read mapping and racon for polishing the sequences.
| + | |
| − | Thus, the assembly pipeline changed to the following calls:
| + | |
| | </p> | | </p> |
| | | | |
| − | <p> | + | <p>As the results indicate, the main contaminant in chloroform phenole extratction is <i>E. coli</i>, as the origin for the native phage DNA. DNAseI and the Norgene kit led to reduction of <i>E. coli</i> contamination. |
| − | MAY BE COLLAPSIBLE
| + | </p> |
| − | #!/usr/bin/env sh
| + | |
| | | | |
| − | INREADS=$1
| + | </div> |
| − | ASMFOLDER=$2
| + | <div class="col-12 col-md-6"> |
| − | ASMPREFIX=$3
| + | |
| | | | |
| − | THREADS=$4
| + | <figure class="figure"> |
| | + | <img src=" https://static.igem.org/mediawiki/2018/1/16/T--Munich--contamination_file_seq.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> Illustration of DNA composition after purification, 3S phage DNA was purified from the <i>E. coli</i> host by chloroform-phenol (CM) and column extraction with and without DNAseI treatment after cell lysis.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | if [ -z "$4" ]
| |
| − | then
| |
| − | THREADS=4
| |
| − | fi
| |
| | | | |
| − | # path to used executables
| |
| − | MINIMAP2=minimap2
| |
| − | MINIASM=miniasm
| |
| − | GRAPHMAP=graphmap
| |
| − | RACON=racon
| |
| | | | |
| − | # first we must overlap all reads with each other
| + | <h3> |
| − | $MINIMAP2 -x ava-ont -t$THREADS $INREADS $INREADS > $ASMFOLDER/$ASMPREFIX.paf
| + | Sequ-Into Allows For Accurate Detection of DNA Contamination Levels |
| | + | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <table class="table"> |
| | + | <p> |
| | + | The laboratory team sequenced T7, 3S, NES and FFP bacteriophages genomes (preparation <a href="https://2018.igem.org/Team:Munich/Protocols">protocols)</a>. |
| | + | </p> |
| | + | <thead> |
| | + | <tr> |
| | + | <th scope="col">Experiment</th> |
| | + | <th scope="col">Sequencing Time</th> |
| | + | <th scope="col">Reads Sequenced</th> |
| | + | <th scope="col">Base-pairs sequenced</th> |
| | + | </tr> |
| | + | </thead> |
| | + | <tbody> |
| | + | <tr> |
| | + | <th scope="row">T7</th> |
| | + | <td>12h 02min</td> |
| | + | <td>424,198</td> |
| | + | <td>1.27 × 10<sup>9</sup></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <th scope="row">3S</th> |
| | + | <td>3h 42min</td> |
| | + | <td>77,092</td> |
| | + | <td>2.53 × 10<sup>8</sup></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <th scope="row">NES</th> |
| | + | <td>3h 58min</td> |
| | + | <td>39,501</td> |
| | + | <td>2.31 × 10<sup>8</sup></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <th scope="row">NFFP</th> |
| | + | <td>5h 42min</td> |
| | + | <td>27,633</td> |
| | + | <td>1.23 × 10<sup>8</sup></td> |
| | + | </tr> |
| | + | </tbody> |
| | + | </table> |
| | | | |
| − | # then miniasm can create alignment
| + | <p> |
| − | $MINIASM -f $INREADS $ASMFOLDER/$ASMPREFIX.paf > $ASMFOLDER/$ASMPREFIX.gfa
| + | The main task after receiving genomes was to assemble the phage genomes from the Nanopore reads without the reference |
| | + | genome and gain the origin sequence. There are several tools available, but for minION Nanopore recommended tools are |
| | + | canu and miniasm, of which we finally used the latter. |
| | + | </p> |
| | + | <p> |
| | + | Despite an excellent lab team we have been confronted with a major problem: contamination. Contamination is a problem |
| | + | regarding the therapeutic usage of the final phage extract, but also an important factor in the bioinformatics pipeline |
| | + | because contamination-based reads can confuse the assembly process. We thus developed <a href="https://2018.igem.org/Team:Munich/Software">sequ-into</a> to detect |
| | + | the contamination and also get rid of contamination-originated reads. In the context of our project, several DNA |
| | + | purification protocols were evaluated with sequ-into and allowed iterative engineering cycles in Phactory, leading to |
| | + | unparalleled purity of up to 96% (bases sequenced). |
| | + | </p> |
| | + | <p> |
| | + | While evaluating the sequencing data with sequ-into we noticed that in the first 10% of the sequencing time of |
| | + | each experiment, more non-target reads were sequenced than in any latter time interval. |
| | + | Moreover we found that this also holds true for the first x sequenced reads, which allowed us to setup sequ-into such |
| | + | that it only analyses the first 1000 reads of a minION sequencing experiment, speeding up computation and enabling an |
| | + | en-run analysis. |
| | + | </p> |
| | | | |
| − | # extract unitigs from miniasm
| + | <div class="row"> |
| − | awk '$1 ~/S/ {print ">"$2"\n"$3}' $ASMFOLDER/$ASMPREFIX.gfa > $ASMFOLDER/$ASMPREFIX.unitigs.fasta
| + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/c/c3/T--munich--time_timeline.png" class="figure-img img-fluid rounded"> |
| | + | <figcaption class="figure-caption">More non-target reads sequenced in the first 10% of the sequencing time of each experiment.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/4/44/T--munich--rank_timeline.png" class="figure-img img-fluid rounded"> |
| | + | <figcaption class="figure-caption">Also in the first x sequenced reads.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| − | # align reads with unitigs
| + | <p> |
| − | $MINIMAP2 $ASMFOLDER/$ASMPREFIX.unitigs.fasta $INREADS > $ASMFOLDER/$ASMPREFIX.unitigs.paf
| + | After getting rid of the contamination we noticed a non-uniform coverage in the phage genome assemblies after |
| | + | re-aligning the reads to the assembly. |
| | + | </p><p> |
| | + | Rearranging the middle part which initially was “over-expressed” and re-assembling led us to the expected uniform |
| | + | coverage of the de-novo genome sequence after re-aligning the input reads. |
| | + | Finally, we could predict coding-sequences and visualize all descriptive statistics in genome diagrams. |
| | + | </p> |
| | | | |
| − | # find contigs from unitigs
| + | </div> |
| − | $RACON $INREADS $ASMFOLDER/$ASMPREFIX.unitigs.paf $ASMFOLDER/$ASMPREFIX.unitigs.fasta > $ASMFOLDER/$ASMPREFIX.contigs.fasta
| + | </div> |
| | | | |
| − | ~/progs/minimap2/minimap2 -x map-ont -a -t$THREADS $ASMFOLDER/$ASMPREFIX.contigs.fasta $INREADS > $ASMFOLDER/$ASMPREFIX.reads.mm2.sam
| |
| − | $GRAPHMAP align -r $ASMFOLDER/$ASMPREFIX.contigs.fasta -d $INREADS -o $ASMFOLDER/$ASMPREFIX.reads.gm.sam
| |
| − | </p>
| |
| | | | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/6/6a/T--Munich--Results_3S_edit_phage.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">3S</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/9/99/T--Munich--results_FFP_edit_phage.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">FFP</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/8/80/T--Munich--Results_NES_phage.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">NES</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | <div></div> |
| | + | <p>More details at <a href="https://2018.igem.org/Team:Munich/DataAnalysis">Data Analysis</a> page.</p> |
| | + | |
| | + | |
| | + | |
| | + | <h2> |
| | + | Encapsulation |
| | + | </h2> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| − | And can be started simply from the command-line using: ./assemble.sh <FQ file> <PATH TO ASM FOLDER> <PREFIX of output>
| + | After successful bacteriophage production and toxicity evaluation the final step of the manufacturing pipeline is the application. |
| − | This finally led to a good assembly after rearranging the middle part which initially was “over-expressed”.
| + | While bacteriophages possess a relatively short have-life when orally administered. The encapsulation in alginate protects phages from the gastric fluid. |
| | + | (Quelle: Colom, Joan, et al. "Microencapsulation with alginate/CaCO 3: A strategy for improved phage therapy." Scientific reports 7 (2017): 41441.), |
| | </p> | | </p> |
| − | <h3>
| |
| − | Proposition of new bacteriophage genomes
| |
| − | </h3>
| |
| | <p> | | <p> |
| − | After having a core genome we want to check how many protein-coding genes we can find on the genome.
| + | We constructed a simple <a href="https://2018.igem.org/Team:Munich/Hardware">device</a> that allows us to encapsulate our bacteriophages in alginate droplets. |
| − | For this task, again several programs exist. Two of the more common programs are glimmer [7] and genemark [8].
| + | |
| − | Because the reputation among the target audience is higher for genemark [9] we used this tool for the genome annotation.
| + | |
| − | We ran the tool on the assembled genome in FASTA format generating a gene annotation file (gff3) for the genome highlighting all coding sequences.
| + | |
| − | For easier and more compact usage, we transformed the genome in fasta format with the annotation in gff3 into the embl flat file format.
| + | |
| − | Finally, we can describe the assembled genomes as follows:
| + | |
| − | TABLE
| + | |
| | </p> | | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| | + | <h3>Droplets are Monodisperse</h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/8/84/T--Munich--Results_brightfield_droplet2_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Alginate composition of 1.8 % Alginate and 0.2 % low-viscosity Alginate delivers spherical Alginate microspheres.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/9/9c/T--Munich--Results_Droplets_zstack-400_8bit.gif" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Alginate encapsulated phages, DNA stained with SYBR Gold.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-4"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/c/c8/T--Munich--Results_brightfield_droplet_disperse_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Monodisperse Alginate Droplets at 150X magnification.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | <p> | | <p> |
| − | Using the embl flat file format we visualized the phage genomes in a circular genome diagram plot (Figure 5).
| + | In order to achieve defined phage concentrations and therefore defined doses, we optimized the monodispersity of our alginate droplets. |
| − | *-+-+*
| + | |
| − | figures
| + | |
| − | *_*_*
| + | |
| − | Here we can see see several things.
| + | |
| − | For the 3S genome, we can see that at certain position we see a high decrease in the coverage (at 58kbp, 72kbp and 110kbp).
| + | |
| − | At these positions no reads align to the reference genome.
| + | |
| − | | + | |
| − | For the NES genome we can see a similar behaviour at 330kbp. Additionally we can see two spikes at the ends of the genome. However, we could also see …
| + | |
| − | Finally the FFP genome again has the same problems as the NES genome ends.
| + | |
| − | Some final bliblubb …
| + | |
| − | | + | |
| | </p> | | </p> |
| | + | <p> |
| | + | In our initial attempts to create alginate droplets the size within a batch often varied significantly. Additionally, due to |
| | + | aggregation a lot of droplets were lost. Optimization of parameters such as flow rate, |
| | + | alginate concentration and N2 pressure led to an increase of monodispersity for all tested sizes (50-300 μm). |
| | + | Specifically, an alginate concentration of 1.8 % alginate and 0.2 % low-viscosity alginate proved to be ideal. Pressure and flow rate determine the droplet sizes. |
| | + | </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/3/38/T--Munich--Results_droplet_optimization.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Monodispersity of alginate droplets with three different diameters before and after optimization of the hardware device.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | <hr> |
| | | | |
| − |
| |
| − | <h2>
| |
| − | Modular Bacteriophage Composition
| |
| − | </h2>
| |
| − | <h2>
| |
| − | Isolation
| |
| − | </h2>
| |
| − | <h3>
| |
| − | Phenol-Chloroform Precipitation Achieves High Titers
| |
| − | </h3>
| |
| − | <h2>
| |
| − | Encapsulation
| |
| − | </h2>
| |
| | <h3> | | <h3> |
| | Bacteriophages Encapsulated In Alginate Can Withstand Gastric Acid | | Bacteriophages Encapsulated In Alginate Can Withstand Gastric Acid |
| | </h3> | | </h3> |
| − |
| |
| − |
| |
| − |
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p>The main problem of oral application is the acidic environment in the gastric fluid, necessitating protective measures against degradation. The other requirement of phage protection is the release of functional phages in the intestines. For this reason we compared the behavior of the encapsulated phages and non-encapsulated phages in simulated gastric fluid (SGF) and simulated intenstinal fluid (SIF). |
| | + | </p> |
| | + | <p> |
| | + | In SGF, the number of active non-encapsulated phages decreases by more than 99.99 % within an hour. This shows the urgent need of a form of protection against degradation to make oral application of bacteriophages possible. As a reference, we used phages that were chemically released by citrate from alginate droplets. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 408: |
Line 1,157: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/4/4f/T--Munich--Results_Joe_SGF_Phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/4/4f/T--Munich--Results_Joe_SGF_Phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">Non-encapsulated phages are highly degraded within an hour in simulated gastric fluid.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| − | <hr>
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p> |
| | + | In comparison, the encapsulated phages were tested in SGF for the same time as the non-encapsulated phages. Afterwards, the same droplets were exposed for two hours to simulated intestinal fluid to test the release of functional bacteriophages in this environment. |
| | + | </p> |
| | + | <p>The encapsulated phages were barely released in an hour of exposure to SGF. After transfering the capsules to SIF the number of active phages reached that of the undegraded reference. |
| | + | This indicates that the encapsulation of bacteriophages in alginate capsules enables the possibility of an oral application. Further experiments could test the alginate capsules in an animal model system. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 423: |
Line 1,174: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/a/a2/T--Munich--Results_Droplets_phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/a/a2/T--Munich--Results_Droplets_phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">A small amount of encapsulated phages are released in SGF. In SIF, functional phages are released in high concentration.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 429: |
Line 1,180: |
| | | | |
| | | | |
| − | | + | <!--- |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | <p> | | <p> |
| | Phactory yields phages with toxicity levels that allow oral administration to the patient. However, oral delivery requires protection of the phages from rapid degradation in the acidic gastric juice, while direct intravenous application requires additional purification steps. To overcome these hurdles, we prototyped two 3D-printed fluidic devices that can be assembled for less than $5. For oral application, we built a nozzle to encapsulate the phages in monodisperse calcium-alginate microspheres protecting them in the stomach. The alginate solution, ejected from a dispenser needle with a syringe pump, was sheared off by a parallel stream of air. Our results show that after 1 hour incubation in simulated gastric fluid, active phages are successfully released in simulated intestinal fluid. For intravenous administration, we can purify the bacteriophages from the remaining cell-extract via fractionation in a pressure-driven size-exclusion filter system. Additionally, we built microfluidic hardware for our human practice project OraColi. </p> | | Phactory yields phages with toxicity levels that allow oral administration to the patient. However, oral delivery requires protection of the phages from rapid degradation in the acidic gastric juice, while direct intravenous application requires additional purification steps. To overcome these hurdles, we prototyped two 3D-printed fluidic devices that can be assembled for less than $5. For oral application, we built a nozzle to encapsulate the phages in monodisperse calcium-alginate microspheres protecting them in the stomach. The alginate solution, ejected from a dispenser needle with a syringe pump, was sheared off by a parallel stream of air. Our results show that after 1 hour incubation in simulated gastric fluid, active phages are successfully released in simulated intestinal fluid. For intravenous administration, we can purify the bacteriophages from the remaining cell-extract via fractionation in a pressure-driven size-exclusion filter system. Additionally, we built microfluidic hardware for our human practice project OraColi. </p> |
| Line 443: |
Line 1,196: |
| | 6. Verhalten im Darm bzw. pH 7 und Pankreatin (2h @ 37°C) ->Phagen relase über Zeit (plot) | | 6. Verhalten im Darm bzw. pH 7 und Pankreatin (2h @ 37°C) ->Phagen relase über Zeit (plot) |
| | </p> | | </p> |
| | + | </div> |
| | + | </div> |
| | + | ---> |
| | + | |
| | + | <hr> |
| | | | |
| − |
| |
| | <div id="phareferences" class="row"> | | <div id="phareferences" class="row"> |
| − | <h2>References</h2> | + | <h2>References [FIX THIS!]</h2> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ol> | | <ol> |
| − | <li> <a href=https://www.biorxiv.org/content/early/2016/05/21/054742 / http://science.sciencemag.org/content/353/6299/aaf5573>Abudayyeh, O.O. et al., 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573.</a></li>
| |
| − | <li><a href=http://www.sciencedirect.com/science/article/pii/S1097276517301338 >Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., Rebecchi, L., Pielak, G.J., Koshland, D., and Goldstein, B., 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65, 975ñ984.</a></li>
| |
| − | <li><a href=https://www.cdc.gov/drugresistance/index.html> Centers for Disease Control and Prevention, 2017. Antibiotic/Antimicrobial Resistance.</a></li>
| |
| − | <li><a href=http://www.gddiergezondheid.nl/diergezondheid/dierziekten/mastitis>GDDiergezondheid, 2017. Mastitis (uierontsteking).</a></li>
| |
| − | <li><a href=http://www.sciencemag.org/lookup/doi/10.1126/science.aam9321> Gootenberg, J.S. et al., 2017. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438ñ442. </a></li>
| |
| − | <li><a href=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5210516/ >Jia, B. et al., 2017. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research 45(D1), D566ñD573.</a></li>
| |
| − | <li><a href="http://science.sciencemag.org/content/337/6096/816.long">Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., Charpentier, E., 2012. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816-821.</a></li>
| |
| − | <li> <a href=http://dx.doi.org/10.1016/j.cell.2017.06.050> Liu, L. et al., 2017. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714ñ726.e10.</a></li>
| |
| − | <li><a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3697360/">McArthur, A.G. et al., 2013. The comprehensive antibiotic resistance database. Antimicrobial Agents and Chemotherapy 57, 3348ñ3357.</a></li>
| |
| − | <li><a href=http://dx.doi.org/10.1016/j.mib.2015.07.004> McArthur, A.G. and Wright, G.D., 2015. Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Current Opinion in Microbiology 27, 45ñ50.</a></li>
| |
| − | <li><a href="https://www.nature.com/articles/nrmicro3525">Schwechheimer, C., and Kuehn, M.J., 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nature Reviews: Microbiology 13, 605-619.</a></li>
| |
| − | <li><a href="https://www.nature.com/articles/s41598-017-05796-x">Sloan, D., Batista, A., and Loeb, A., 2017. The Resilience of Life to Astrophysical Events. Scientific Reports 7, 5419-5424.</a></li>
| |
| − | <li><a href=http://www.zuivelnl.org/wp-content/uploads/2016/09/Dutch-dairy-in-figures-2015.pdf> Statistics Netherlands, 2015. Dutch dairy in figures. </a></li>
| |
| − | <li><a href=http://www.who.int/mediacentre/factsheets/antibiotic-resistance/en> World Health Organization, 2016. Antibiotic resistance. </a> </li>
| |
| | <li><a href="http://www.sciencedirect.com/science/article/pii/S0092867415012003?via%3Dihub">Zetsche, B, Gootenberg, J.S., Abudayyeh, O.O., Slaymaker, I.M., Makarova, K.S., Essletzbichler, P., Volz, S.E., van der Oost, J., Regev, Aviv, Koonin, E.V., Zhang, F., 2015. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759-771. </a></li> | | <li><a href="http://www.sciencedirect.com/science/article/pii/S0092867415012003?via%3Dihub">Zetsche, B, Gootenberg, J.S., Abudayyeh, O.O., Slaymaker, I.M., Makarova, K.S., Essletzbichler, P., Volz, S.E., van der Oost, J., Regev, Aviv, Koonin, E.V., Zhang, F., 2015. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759-771. </a></li> |
| | </ol> | | </ol> |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| + | |
| − |
| + | |
| | | | |
| | </main> | | </main> |