| (11 intermediate revisions by 4 users not shown) | |||
| Line 35: | Line 35: | ||
| − | |||
| − | <h2><span class="parts-color"> | + | <h2><span class="parts-color">Silver medal criterion #1 </span></h2> |
<p style="font-size=120%;"> | <p style="font-size=120%;"> | ||
| − | + | <p><b><center>How many cells do actually attach in the biofilm?</b></center</p> | |
| − | + | <p> | |
| − | + | Not all bacteria will attach in a biofilm, and we thought it would be interesting to get an indication about the ratio between cells attached in biofilm and that for planktonic bacteria. As an indicator, a fluorescence protein called RRvT was used and characterized to show the growth of DH5a in LB medium. This was compared to the absorbance-values of DH5a cells attached in biofilm, analyzed by a Crystal Violet assay. For the DH5a cells attached in biofilm they had even grown on 0.4% glucose, but the absorbance values for the planktonic bacteria is still significantly higher than for the attached bacteria. We had also discovered from articles that RRvT works better than RFP in acidic conditions. RRvT actually has a pkA of 3.9. The thought was that if the bacteria needed to grow in acidic conditions in a biofilm, RRvT could still be used to compare the growth in biofilm with the growth of planktonic bacteria. RRvT works as expected because the absorption increases with the growth of the bacteria, but we also discovered some limitations to this method over time. The wells gets saturated with RRvT at a point where the absorption values will not change significantly. </p> | |
| − | The | + | |
| − | < | + | |
<br> | <br> | ||
| − | + | <br><p> | |
| − | + | To change a previous part, we decided to change the sequence of the red fluorescent protein coding device BBa_J04450. This was done by replacing the RFP sequence with the sequence for the RRvT. RRvT is a red fluorescent protein with a much higher brightness, and it can be used when the concentration of the inducer is to low to activate the BBa_J04450 biobrick. </p> | |
| − | + | ||
<br> | <br> | ||
| + | <br><p> | ||
| + | The changed biobrick was synthesized by IDT as a linear sequence containing prefix and suffix. The prefix and suffix sequences can be found at parts.igem.org, and contain restriction enzyme cut sites that can be used to transfer and assemble parts. We then ligated it with the pSB1C3 backbone to make it RFC10 compatible. The changed biobrick contains a lactose induced promoter (Plac BBa_R0010), a ribosome binding site (BBa_B0034), the RRvT sequence and a terminator (BBa_B0015). The new biobrick is therefore a composite part. We have not sent the basic part with the RRvT sequence alone into the registry. Only the composite part was tested and sent in to the iGEM registry.</p> | ||
<br> | <br> | ||
| − | The following figure shows growth curves for RRvT and mRFP as absorbance against time. | + | <br><p> |
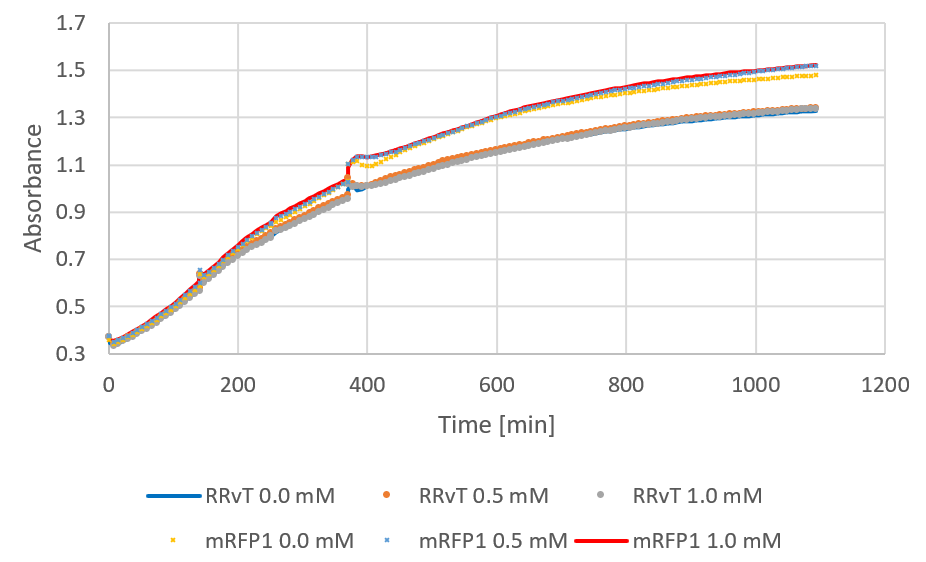
| − | <img src=""> | + | The following figure shows growth curves for RRvT and mRFP as absorbance against time. This was compared to the growth in absorbance for DH5a in LB medium (0.4% glucose) for the biofilm measurements (see results). Both curves showed an increase, and the absorbance values for RRvT (planktonic bacteria) were much higher than for attached bacteria in biofilm compared at the same time points. </p> |
| + | <figure><img src="https://static.igem.org/mediawiki/2018/f/f9/T--NTNU_Trondheim--growth.png"><div><b>Figure 1:</b> Growth curve of RRvT and mRFP1. After around 250 minutes, 0.5 mM IPTG was added.</div></figure> | ||
<br> | <br> | ||
<br> | <br> | ||
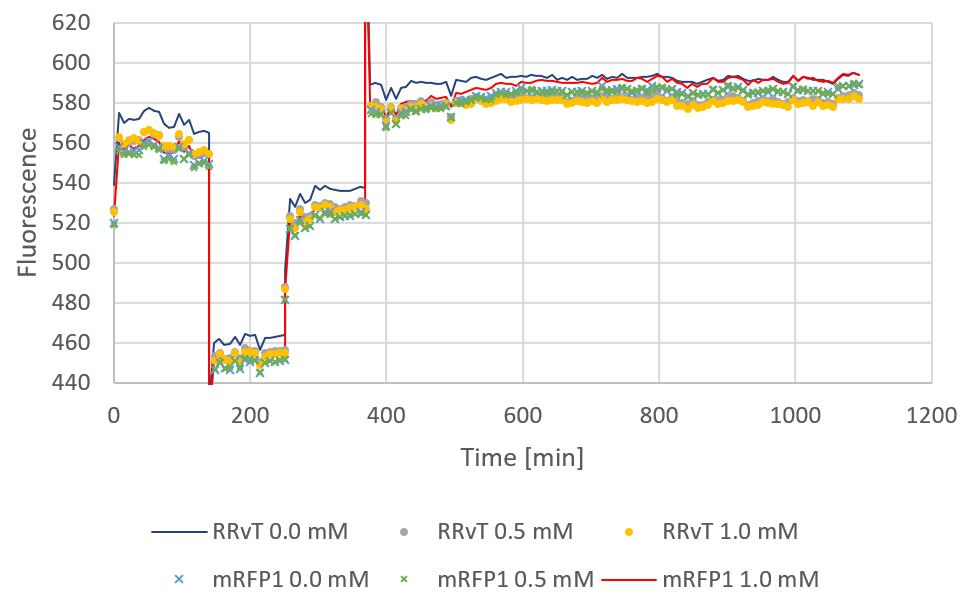
| − | Fluorescence plotted against time for the two biobricks are shown in the figure below:</p> | + | <p>Fluorescence plotted against time for the two biobricks are shown in the figure below:</p> |
<br> | <br> | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/ | + | <img src="https://static.igem.org/mediawiki/2018/1/1d/T--NTNU_Trondheim--blablaba.png"> |
| − | <div><b>Figure | + | <div><b>Figure 2:</b> Fluorescence for both biobricks plotted against time. The discontinuous points of the curve are due to a shut-down of the computer during measurements. 0, 0.5 and 1.0 mM IPTG was added at starting time.</div> |
</figure><br> | </figure><br> | ||
| − | |||
<br> | <br> | ||
Latest revision as of 12:32, 6 November 2018

Silver medal criterion #1
Not all bacteria will attach in a biofilm, and we thought it would be interesting to get an indication about the ratio between cells attached in biofilm and that for planktonic bacteria. As an indicator, a fluorescence protein called RRvT was used and characterized to show the growth of DH5a in LB medium. This was compared to the absorbance-values of DH5a cells attached in biofilm, analyzed by a Crystal Violet assay. For the DH5a cells attached in biofilm they had even grown on 0.4% glucose, but the absorbance values for the planktonic bacteria is still significantly higher than for the attached bacteria. We had also discovered from articles that RRvT works better than RFP in acidic conditions. RRvT actually has a pkA of 3.9. The thought was that if the bacteria needed to grow in acidic conditions in a biofilm, RRvT could still be used to compare the growth in biofilm with the growth of planktonic bacteria. RRvT works as expected because the absorption increases with the growth of the bacteria, but we also discovered some limitations to this method over time. The wells gets saturated with RRvT at a point where the absorption values will not change significantly.
To change a previous part, we decided to change the sequence of the red fluorescent protein coding device BBa_J04450. This was done by replacing the RFP sequence with the sequence for the RRvT. RRvT is a red fluorescent protein with a much higher brightness, and it can be used when the concentration of the inducer is to low to activate the BBa_J04450 biobrick.
The changed biobrick was synthesized by IDT as a linear sequence containing prefix and suffix. The prefix and suffix sequences can be found at parts.igem.org, and contain restriction enzyme cut sites that can be used to transfer and assemble parts. We then ligated it with the pSB1C3 backbone to make it RFC10 compatible. The changed biobrick contains a lactose induced promoter (Plac BBa_R0010), a ribosome binding site (BBa_B0034), the RRvT sequence and a terminator (BBa_B0015). The new biobrick is therefore a composite part. We have not sent the basic part with the RRvT sequence alone into the registry. Only the composite part was tested and sent in to the iGEM registry.
The following figure shows growth curves for RRvT and mRFP as absorbance against time. This was compared to the growth in absorbance for DH5a in LB medium (0.4% glucose) for the biofilm measurements (see results). Both curves showed an increase, and the absorbance values for RRvT (planktonic bacteria) were much higher than for attached bacteria in biofilm compared at the same time points.
Fluorescence plotted against time for the two biobricks are shown in the figure below: