| (22 intermediate revisions by 6 users not shown) | |||
| Line 6: | Line 6: | ||
left: 125px; | left: 125px; | ||
} | } | ||
| − | # | + | #project_small { |
background-color: #292929; | background-color: #292929; | ||
} | } | ||
| Line 27: | Line 27: | ||
} | } | ||
} | } | ||
| − | |||
| − | |||
| − | |||
</style> | </style> | ||
| Line 40: | Line 37: | ||
<div id="index" class="block"> | <div id="index" class="block"> | ||
<div id="indexContent"> | <div id="indexContent"> | ||
| − | + | <p><a href="#Background" class="link">Background</a></p> | |
| − | <p><a href="# | + | <p><a href="#Solution" class="link">Our Solution</a></p> |
<p><a href="#Results" class="link">Results</a></p> | <p><a href="#Results" class="link">Results</a></p> | ||
<p><a href="#References" class="link">References</a></p> | <p><a href="#References" class="link">References</a></p> | ||
| Line 50: | Line 47: | ||
</div> | </div> | ||
<div id="MainContent"> | <div id="MainContent"> | ||
| + | <div class="block title"><h1 id="Background">BACKGROUND</h1></div> | ||
<div class="block full"><p><i>When we first designed our project, it soon came to our mind that we had to ensure the containment of our bacteria from the environment. The best way to do that, apart from the physical containment performed by the <a href="https://2018.igem.org/Team:Pasteur_Paris/Membrane">porous membrane</a>, was to implement our bacterial interface with a kill-switch. </i></p></div> | <div class="block full"><p><i>When we first designed our project, it soon came to our mind that we had to ensure the containment of our bacteria from the environment. The best way to do that, apart from the physical containment performed by the <a href="https://2018.igem.org/Team:Pasteur_Paris/Membrane">porous membrane</a>, was to implement our bacterial interface with a kill-switch. </i></p></div> | ||
| − | |||
<div class="block title"><h3 style="text-align: left;">What is a kill-switch? </h3></div> | <div class="block title"><h3 style="text-align: left;">What is a kill-switch? </h3></div> | ||
<div class="block full"> | <div class="block full"> | ||
| − | <p>A kill-switch is, literally, something that tells a bacterium whether to live or die, depending on specific environmental conditions. | + | <p>A kill-switch is, literally, something that tells a bacterium whether to live or die, depending on specific environmental conditions. With the rapid development of synthetic biology, these switches are of many kinds and have been increasingly used in recent years <sup>[1]</sup>. They mainly work by introducing in the bacteria gene constructs that block important genes or express toxins when exposed to a specific environmental condition. We realized an educational video destined to explain to the public audience the principle of a kill-switch. This video can be found <a href="https://2018.igem.org/Team:Pasteur_Paris/Public_Engagement">here</a>.</p> |
</div> | </div> | ||
| + | <div class="block separator-mark"></div> | ||
| + | <div class="block title"><h1 id="Solution">OUR SOLUTION</h1></div> | ||
<div class="block title"><h3 style="text-align: left;" id="Our">Our kill-switch</h3></div> | <div class="block title"><h3 style="text-align: left;" id="Our">Our kill-switch</h3></div> | ||
<div class="block full"> | <div class="block full"> | ||
| − | <p>Our interface is eventually destined to “cohabit” in a human body. We want to make sure that the bacteria we create cannot develop outside of the space we put | + | <p>Our interface is eventually destined to “cohabit” in a human body. We want to make sure that the bacteria we create cannot develop outside of the space we put them in. Thus, our kill-switch makes sure that when the environmental conditions of the inner human body are not met, the bacteria die. </p> |
| − | <p>Reviewing the literature led us to come across a very interesting article presenting the cryodeath kill-switch, invented by Finn Stirling in 2017, which is triggered by temperatures under 37°C [2]. For obvious reasons, this kill switch sounded like a perfect fit | + | <p>Reviewing the literature led us to come across a very interesting article presenting the cryodeath kill-switch, invented by Finn Stirling in 2017, which is triggered by temperatures under 37°C <sup>[2]</sup>. For obvious reasons, this kill switch sounded like a perfect fit for our application. Contacting Finn comforted us in that choice, and we had the chance to exchange a lot, not only concerning the kill switch but on our project in general. After reviewing the original sequence, we modified it so that it would become a biobrick, and we removed what was unnecessary to us, leading to the making of our biobrick <a href="http://parts.igem.org/Part:BBa_K2616002" style="font-weight: bold ; color:#85196a;" target="__blank">BBa_K2616002.</a></p> |
</div> | </div> | ||
<div class="block title"><h3 style="text-align: left;" id="Modified">The modified cryodeath kill-switch</h3></div> | <div class="block title"><h3 style="text-align: left;" id="Modified">The modified cryodeath kill-switch</h3></div> | ||
<div class="block full"> | <div class="block full"> | ||
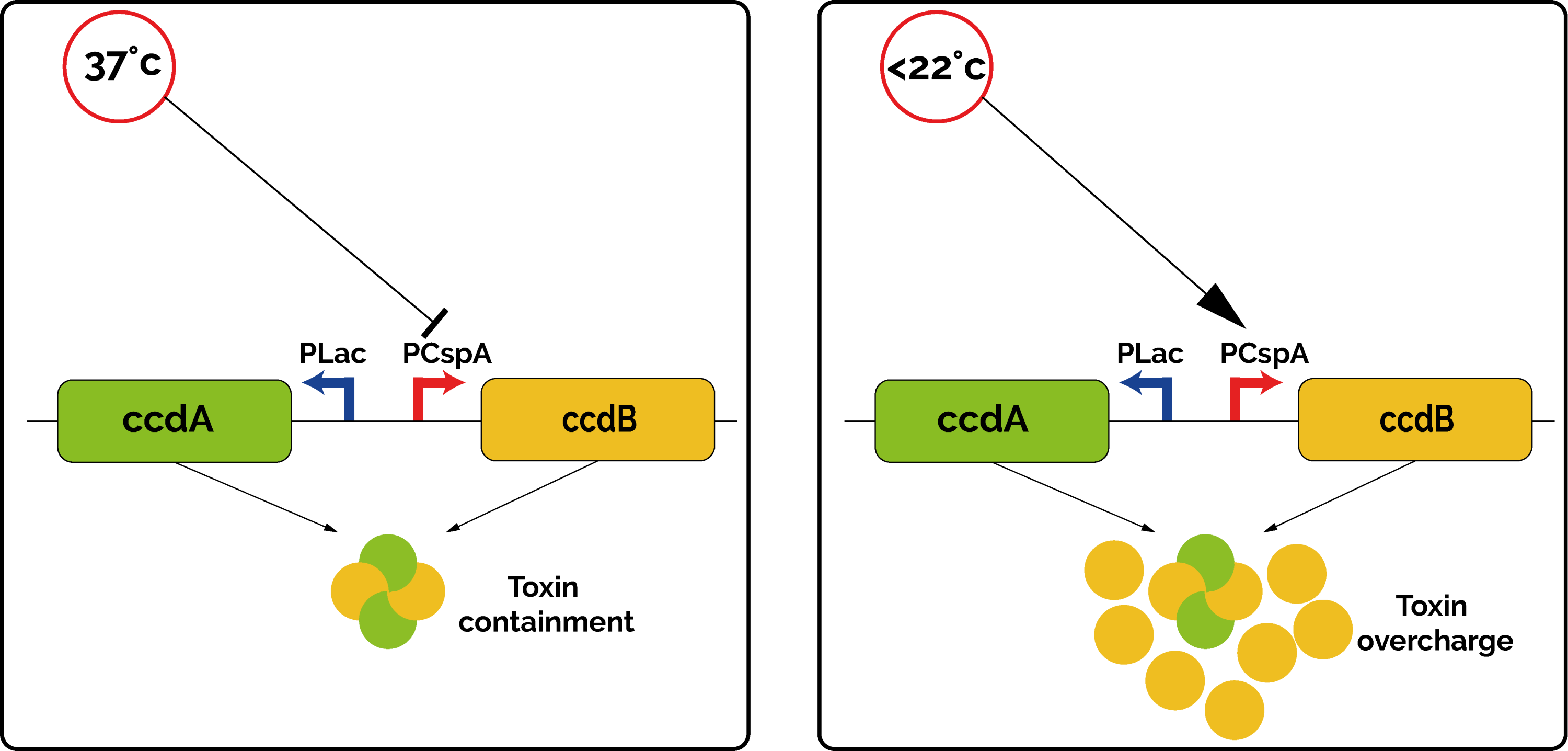
| − | <p>The kill-switch functions by the means of the toxin/antitoxin couple CcdB/CcdA. The toxin targets and inhibits the GyrA subunit of DNA gyrase, an essential bacterial enzyme that catalyzes the super-coiling of double-stranded closed circular DNA [3]. </p> | + | <p>The kill-switch functions by the means of the toxin/antitoxin couple CcdB/CcdA. The toxin targets and inhibits the GyrA subunit of DNA gyrase, an essential bacterial enzyme that catalyzes the super-coiling of double-stranded closed circular DNA <sup>[3]</sup>. </p> |
| − | <p>In the kill-switch, the transcription of CcdB and CcdA are regulated by two different promoters. CcdA is expressed by the constitutive promoter P<sub>lac</sub>, which ensures a constant low production of antitoxin [4]. On the contrary, the toxin CcdB is expressed by the promoter PcspA. This promoter was originally discovered upstream of the Cold Shock Protein A in many bacteria, as a way to cope with environmental stress [5]. This promoter ensures a very low transcription rate at 37°C, but it increases drastically below 22°C. </p> | + | <p>In the kill-switch, the transcription of CcdB and CcdA are regulated by two different promoters. CcdA is expressed by the constitutive promoter P<sub>lac</sub>, which ensures a constant low production of antitoxin <sup>[4]</sup>. On the contrary, the toxin CcdB is expressed by the promoter PcspA. This promoter was originally discovered upstream of the Cold Shock Protein A (CspA) in many bacteria, as a way to cope with environmental stress. Indeed, when the temperature drops below 37°C, the promoter PcspA increases the level of expression of CspA gradually <sup>[5]</sup>. This promoter ensures a very low transcription rate at 37°C, but it increases drastically below 22°C. </p> |
| − | <p>Overall, at 37°C, the quantity of antitoxin CcdA is high enough to cope with the leaky low level of toxin produced. However, if the bacteria happen to be in an environment at a lower temperature, the toxin promoter is not repressed anymore, the quantity of toxin becomes too important, and the bacteria is not able to grow ( | + | <p>Overall, at 37°C, the quantity of antitoxin CcdA is high enough to cope with the leaky low level of toxin produced. However, if the bacteria happen to be in an environment at a lower temperature, the toxin promoter is not repressed anymore, the quantity of toxin becomes too important, and the bacteria is not able to grow (Figure 1). This allows us to make sure that our genetically modified organisms will not spread out in an open environment. </p> |
| + | <br> | ||
<img src="https://static.igem.org/mediawiki/2018/f/fd/T--Pasteur_Paris--kill-switch-functioning.png"> | <img src="https://static.igem.org/mediawiki/2018/f/fd/T--Pasteur_Paris--kill-switch-functioning.png"> | ||
<div class="legend"><b>Figure 1: </b>Functioning of the cryodeath kill-switch at 37°C and at 22°C.</div> | <div class="legend"><b>Figure 1: </b>Functioning of the cryodeath kill-switch at 37°C and at 22°C.</div> | ||
</div> | </div> | ||
| + | <div class="block separator-mark"></div> | ||
| − | <div class="block title">< | + | <div class="block title"><h1 id="Results">RESULTS</h1></div> |
<div class="block full"> | <div class="block full"> | ||
| − | + | <i><p>Achievements:<br></i> | |
| − | + | <ul style="text-align: left;list-style: disc;"> | |
| − | + | <li>Successfully cloned the biobrick <a href="http://parts.igem.org/Part:BBa_K2616002"style="font-weight: bold ; color:#85196a;"target="_blank"> Bba_K2616002</a> coding for toxin/antitoxin (CcdB/CcdA) system in pSB1C3, creating a <b>new part</b>.</li> | |
| − | + | <li>Successfully sequenced <a href="http://parts.igem.org/Part:BBa_K2616002"style="font-weight: bold ; color:#85196a;"target="_blank"> BBa_K2616002</a> in pSB1C3 and sent it to iGEM registry.</li> | |
| − | + | <li>Successfully observed <b>normal growth</b> of our engineered bacteria at 25°C and 37°C and <b>absence of growth</b> at 18°C and 20°C, showing the <b>efficiency of the kill switch</b>.</li> | |
| − | + | </ul><br></p> | |
| − | + | <p><i>Next steps:</i><br> | |
| − | + | <ul style="text-align: left;list-style: disc;"> | |
| − | + | <li>Find a system that kills bacteria when released in the environment rather than just stopping their growth.</li> | |
| + | </ul> | ||
| + | </p></p> | ||
</div> | </div> | ||
| + | <div><p style="text-align: center;"> Visit the <a href="https://2018.igem.org/Team:Pasteur_Paris/Results"style="font-weight: bold ; color:#85196a;" target="__blank">RESULTS page </a> to get more information</p></div> | ||
| + | |||
| + | |||
| Line 89: | Line 95: | ||
<div class="block title"><h1 id="References">REFERENCES</h1></div> | <div class="block title"><h1 id="References">REFERENCES</h1></div> | ||
<div class="block full"> | <div class="block full"> | ||
| − | <ul style="text-align: left;"> | + | <ul style="text-align: left;list-style: disc;"> |
<li style="list-style-type: decimal;">“Kill switches limit modified microbes,” Nature, vol. 528, no. 7581, pp. 166–167, Dec. 2015.<br><br></li> | <li style="list-style-type: decimal;">“Kill switches limit modified microbes,” Nature, vol. 528, no. 7581, pp. 166–167, Dec. 2015.<br><br></li> | ||
<li style="list-style-type: decimal;">F. Stirling et al., “Rational Design of Evolutionarily Stable Microbial Kill Switches,” Mol. Cell, vol. 68, no. 4, p. 686–697.e3, Nov. 2017.<br><br></li> | <li style="list-style-type: decimal;">F. Stirling et al., “Rational Design of Evolutionarily Stable Microbial Kill Switches,” Mol. Cell, vol. 68, no. 4, p. 686–697.e3, Nov. 2017.<br><br></li> | ||
<li style="list-style-type: decimal;">R. J. Reece and A. Maxwell, “DNA Gyrase: Structure and Function,” Crit. Rev. Biochem. Mol. Biol., vol. 26, no. 3–4, pp. 335–375, Jan. 1991.<br><br></li> | <li style="list-style-type: decimal;">R. J. Reece and A. Maxwell, “DNA Gyrase: Structure and Function,” Crit. Rev. Biochem. Mol. Biol., vol. 26, no. 3–4, pp. 335–375, Jan. 1991.<br><br></li> | ||
| − | <li style="list-style-type: decimal;">T. P. Malan and W. R. McClure, “Dual promoter control of the <i> | + | <li style="list-style-type: decimal;">T. P. Malan and W. R. McClure, “Dual promoter control of the <i> E. coli </i> lactose operon.,” Cell, vol. 39, no. 1, pp. 173–80, Nov. 1984.<br><br></li> |
<li style="list-style-type: decimal;">R. Keto-Timonen, N. Hietala, E. Palonen, A. Hakakorpi, M. Lindström, and H. Korkeala, “Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia,” Front. Microbiol., vol. 7, Jul. 2016.<br><br></li> | <li style="list-style-type: decimal;">R. Keto-Timonen, N. Hietala, E. Palonen, A. Hakakorpi, M. Lindström, and H. Korkeala, “Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia,” Front. Microbiol., vol. 7, Jul. 2016.<br><br></li> | ||
Latest revision as of 14:50, 10 November 2018
BACKGROUND
When we first designed our project, it soon came to our mind that we had to ensure the containment of our bacteria from the environment. The best way to do that, apart from the physical containment performed by the porous membrane, was to implement our bacterial interface with a kill-switch.
What is a kill-switch?
A kill-switch is, literally, something that tells a bacterium whether to live or die, depending on specific environmental conditions. With the rapid development of synthetic biology, these switches are of many kinds and have been increasingly used in recent years [1]. They mainly work by introducing in the bacteria gene constructs that block important genes or express toxins when exposed to a specific environmental condition. We realized an educational video destined to explain to the public audience the principle of a kill-switch. This video can be found here.
OUR SOLUTION
Our kill-switch
Our interface is eventually destined to “cohabit” in a human body. We want to make sure that the bacteria we create cannot develop outside of the space we put them in. Thus, our kill-switch makes sure that when the environmental conditions of the inner human body are not met, the bacteria die.
Reviewing the literature led us to come across a very interesting article presenting the cryodeath kill-switch, invented by Finn Stirling in 2017, which is triggered by temperatures under 37°C [2]. For obvious reasons, this kill switch sounded like a perfect fit for our application. Contacting Finn comforted us in that choice, and we had the chance to exchange a lot, not only concerning the kill switch but on our project in general. After reviewing the original sequence, we modified it so that it would become a biobrick, and we removed what was unnecessary to us, leading to the making of our biobrick BBa_K2616002.
The modified cryodeath kill-switch
The kill-switch functions by the means of the toxin/antitoxin couple CcdB/CcdA. The toxin targets and inhibits the GyrA subunit of DNA gyrase, an essential bacterial enzyme that catalyzes the super-coiling of double-stranded closed circular DNA [3].
In the kill-switch, the transcription of CcdB and CcdA are regulated by two different promoters. CcdA is expressed by the constitutive promoter Plac, which ensures a constant low production of antitoxin [4]. On the contrary, the toxin CcdB is expressed by the promoter PcspA. This promoter was originally discovered upstream of the Cold Shock Protein A (CspA) in many bacteria, as a way to cope with environmental stress. Indeed, when the temperature drops below 37°C, the promoter PcspA increases the level of expression of CspA gradually [5]. This promoter ensures a very low transcription rate at 37°C, but it increases drastically below 22°C.
Overall, at 37°C, the quantity of antitoxin CcdA is high enough to cope with the leaky low level of toxin produced. However, if the bacteria happen to be in an environment at a lower temperature, the toxin promoter is not repressed anymore, the quantity of toxin becomes too important, and the bacteria is not able to grow (Figure 1). This allows us to make sure that our genetically modified organisms will not spread out in an open environment.
RESULTS
Achievements:
- Successfully cloned the biobrick Bba_K2616002 coding for toxin/antitoxin (CcdB/CcdA) system in pSB1C3, creating a new part.
- Successfully sequenced BBa_K2616002 in pSB1C3 and sent it to iGEM registry.
- Successfully observed normal growth of our engineered bacteria at 25°C and 37°C and absence of growth at 18°C and 20°C, showing the efficiency of the kill switch.
Next steps:
- Find a system that kills bacteria when released in the environment rather than just stopping their growth.
Visit the RESULTS page to get more information
REFERENCES
- “Kill switches limit modified microbes,” Nature, vol. 528, no. 7581, pp. 166–167, Dec. 2015.
- F. Stirling et al., “Rational Design of Evolutionarily Stable Microbial Kill Switches,” Mol. Cell, vol. 68, no. 4, p. 686–697.e3, Nov. 2017.
- R. J. Reece and A. Maxwell, “DNA Gyrase: Structure and Function,” Crit. Rev. Biochem. Mol. Biol., vol. 26, no. 3–4, pp. 335–375, Jan. 1991.
- T. P. Malan and W. R. McClure, “Dual promoter control of the E. coli lactose operon.,” Cell, vol. 39, no. 1, pp. 173–80, Nov. 1984.
- R. Keto-Timonen, N. Hietala, E. Palonen, A. Hakakorpi, M. Lindström, and H. Korkeala, “Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia,” Front. Microbiol., vol. 7, Jul. 2016.

