|
|
| (25 intermediate revisions by 3 users not shown) |
| Line 4: |
Line 4: |
| | | | |
| | .pictureTitle{ | | .pictureTitle{ |
| − | background: linear-gradient(rgba(0,0,0,.5), rgba(0,0,0,.8)), url("https://static.igem.org/mediawiki/2018/b/bf/T--Munich--background_results.png"); | + | background: linear-gradient(rgba(0,0,0,.3), rgba(0,0,0,.5)), url("https://static.igem.org/mediawiki/2018/6/66/T--Munich--header_applied_design.png"); |
| | background-repeat: no-repeat; | | background-repeat: no-repeat; |
| | background-size: cover; | | background-size: cover; |
| Line 28: |
Line 28: |
| | font-size: 1.5rem; | | font-size: 1.5rem; |
| | } | | } |
| − |
| |
| | | | |
| | @media only screen and (max-width: 809.99px) { | | @media only screen and (max-width: 809.99px) { |
| Line 73: |
Line 72: |
| | </div> | | </div> |
| | ---> | | ---> |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | Phages are bacteria specific organisms, not harmful to humans and more specific than antibiotics. With Phactory it is possible to produce therapeutically relevant phages independent of the corresponding pathogen. This was done in our optimized home-made cell extract. With Phactory, we achieved a stable manufacturing process which produces phages with the titer and purity required for patient treatment. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| | <h2>Optimizing a Cell-Free Expression System</h2> | | <h2>Optimizing a Cell-Free Expression System</h2> |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | The first part of our project is the optimization of a cell-free expression system as a manufacturing platform for bacteriophages. For our purpose, it is necessary to produce a high quality cell extract, in a reproducible and easy manner. | + | The first part of our project is the optimization of a cell-free expression system as a manufacturing platform for bacteriophages. |
| − | <br>
| + | For our purpose, it is necessary to produce a high-quality cell extract, in a reproducible and easy manner. |
| | We focused on achieving the following goals for our cell extract: | | We focused on achieving the following goals for our cell extract: |
| | </p> | | </p> |
| Line 85: |
Line 91: |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ul style=""> | | <ul style=""> |
| − | <li>increase protein content</li> | + | <li>increasing the protein content</li> |
| − | <li>find reproducible methods of quality control</li> | + | <li>finding reproducible methods of quality control</li> |
| − | <li>produce cell-extract that allows phage assembly</li> | + | <li>producing cell-extract that allows phage assembly</li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| | <br> | | <br> |
| − | Considering a possible commercial use of our product we decided to furthermore evaluate the potential for scaling up of our preparation protocol. | + | Considering a possible commercial application of our product we decided to furthermore evaluate the potential for scaling up of our preparation protocol. |
| | <br> | | <br> |
| − | We chose to try several different approaches to achieve these goals | + | We chose to try several different approaches to achieve these goals: |
| | </p> | | </p> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ul style=""> | | <ul style=""> |
| − | <li>test cell cultivation in a bioreactor to enable upscaling</li> | + | <li>testing cell cultivation in a bioreactor to enable upscaling</li> |
| − | <li>find optimal lysis conditions that produce high-quality extract</li> | + | <li>finding optimal lysis conditions that produce high-quality extract</li> |
| − | <li>test scaling up of cell lysis</li> | + | <li>scaling up of cell lysis</li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| Line 125: |
Line 131: |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ul style=""> | | <ul style=""> |
| | <li>bioreactor cultivation allows for upscaling of cell extract production</li> | | <li>bioreactor cultivation allows for upscaling of cell extract production</li> |
| | <li>sonication and lysozyme improve the performance of cell extract</li> | | <li>sonication and lysozyme improve the performance of cell extract</li> |
| − | <li>cultivation and extract preparation barely impacts cell extract composition </li> | + | <li>cultivation and extract preparation barely impact cell extract composition </li> |
| | <li>lyophilization is a good choice for cell-extract storage</li> | | <li>lyophilization is a good choice for cell-extract storage</li> |
| | </ul> | | </ul> |
| Line 141: |
Line 147: |
| | <li>biorector cultivation, sonication, lysozyme enhance the performance</li> | | <li>biorector cultivation, sonication, lysozyme enhance the performance</li> |
| | <li>different cultivation and cell disruption methods do not overly impact the protein composition, but probably the activity of the expressed protein</li> | | <li>different cultivation and cell disruption methods do not overly impact the protein composition, but probably the activity of the expressed protein</li> |
| − | <li>lyophylization is a good way to store cell extract</li> | + | <li>lyophilization is a good way to store cell extract</li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| Line 156: |
Line 162: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Cultivating bacteria is the first step in cell extract preparation. The cell extract preparation protocol of Sun et al. uses shaking flask cultivation for biomass production. It recommends cell harvest in the mid-log growth phase at OD 1.8-2.0 to produce high-quality cell extract. We recorded growth curves for shaking flask cultivation and bioreactor cultivation in a lab-scale bioreactor, to compare biomass production under different culture conditions. | + | <p>Cultivating bacteria is the first step in cell extract preparation. The cell extract preparation protocol of Sun et al.<sup><a href="#phareferences">1</a></sup> uses shaking flask cultivation for biomass production. It recommends cell harvest in the mid-log growth phase at OD 1.8-2.0 to produce high-quality cell extract. We recorded growth curves for shaking flask cultivation and bioreactor cultivation in a lab-scale bioreactor, to compare biomass production under different culture conditions. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 169: |
Line 175: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>The growth curve from shaking flask cultivation, revealed that the mid-log growth phase correlates to an OD of around 2. In bioreactor cultivation the mid-log growth phase is prolonged, due to the better aeration of the medium, and corresponds to an OD between 4 and 6. We harvested cells at OD 4, 5 and 6 to test which harvest point is most suitable to produce high quality cell extract. | + | <p>The growth curve from shaking flask cultivation revealed that the mid-log growth phase correlates to an OD of around 2. In bioreactor cultivation the mid-log growth phase is prolonged, due to the better aeration of the medium, and corresponds to an OD between 4 and 6. We harvested cells at OD 4, 5 and 6 to test which harvest point is most suitable to produce high quality cell extract. |
| | </p> | | </p> |
| | | | |
| Line 184: |
Line 190: |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | The test reveals that the protein content is higher when cells where harvested at higher OD, with OD 5 yielding the highest protein content. In addition, the cell extract from OD 5 produces the highest fluorescence intensity in the TXTL test. Compared to a cell extract from shaking flask cultivation the signal is reduced to about 60 %. The most likely cause for this result, is that a higher fraction of proteins in the OD 5 cell extract is inactive, possibly due to protein damage during cell lysis (at the time of this experiment, cell lysis was not yet fully optimized). | + | The test revealed that the protein content is higher when cells were harvested at a higher OD, with OD 5 yielding the highest protein content. In addition, the cell extract from OD 5 produces the highest fluorescence intensity in the TXTL test. |
| | + | Compared to a cell extract from shaking flask cultivation the signal is reduced to about 60 %. The most likely cause for this result |
| | + | is that a higher fraction of proteins in the OD 5 cell extract is inactive, possibly due to protein damage during cell lysis (at the time of this experiment, cell lysis was not yet fully optimized). |
| | </p> | | </p> |
| | <p> | | <p> |
| − | The comparison cultivation methods revealed that cultivation in a bioreactor is an applicable alternative to shaking flask cultivation and offers great potential for upscaling of cell extract preparation. In our 2 L lab-scale bioreactor we were able to produce 20 g of cell pellet, compared to 4-5 g that 2 L of shaking flask cultivation yield. | + | The comparison of cultivation methods revealed that cultivation in a bioreactor is an applicable alternative to shaking flask cultivation and offers great potential for upscaling of cell extract preparation. In our 2 L lab-scale bioreactor we were able to produce 20 g of cell pellet, compared to 4-5 g that 2 L of shaking flask cultivation yield. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 200: |
Line 208: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
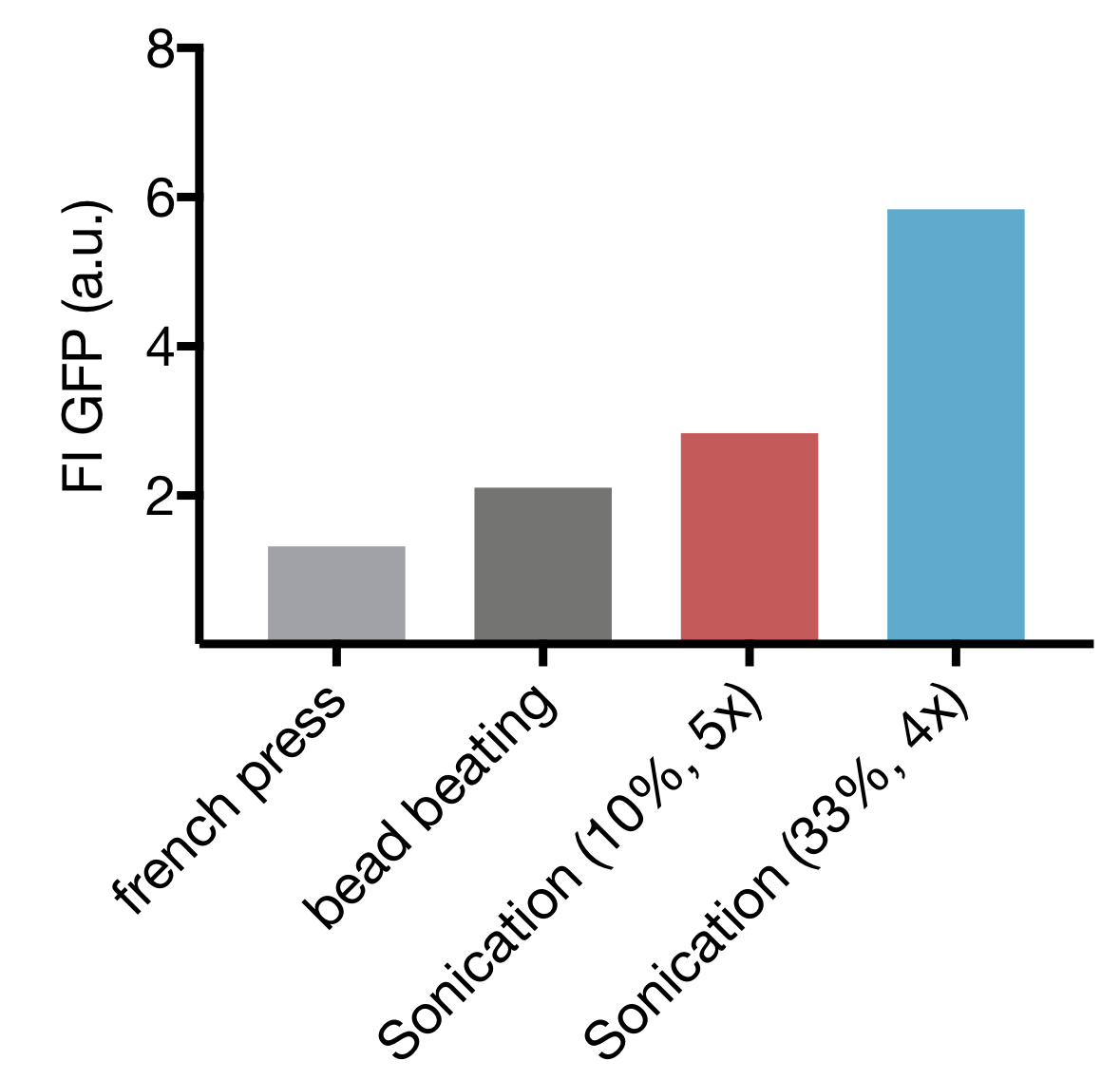
| − | <p>We tested three different cell lysis methods: beat beating, French press and sonication. Beat beating is an inexpensive and widely employed method but upscaling is problematic and the preparation is tedious and time-consuming. High-pressure cell disruption in a so called ‘French-press’ is also used but these devices are very expensive and not widely prevalent. This contradicts our effort to provide a generally-applicable protocol for preparation of home-made cell extract. A sonication device on the other hand is available in most biochemical labs as it is a commonly used method for cell lysis in protein purification protocols. Nevertheless, we started by comparing all three lysis methods with settings commonly applied for cell lysis. | + | <p>We tested three different cell lysis methods: beat beating, French press and sonication. Beat beating is an inexpensive and widely employed method |
| | + | but upscaling is problematic, and the preparation is tedious and time-consuming. High-pressure cell disruption in a so called ‘French-press’ is also |
| | + | used but these devices are very expensive and not widely prevalent. This contradicts our effort to provide a generally-applicable protocol for preparation |
| | + | of home-made cell extract. A sonication device on the other hand is available in most biochemical labs as it is a commonly used method for cell lysis in |
| | + | protein purification protocols. Nevertheless, we started by comparing all three lysis methods with settings commonly applied for cell lysis. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 207: |
Line 219: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Our initial test showed that cell lysis by sonication yields extract with up to 2.8 and 4.4 fold higher expression than cell lysis by bead beating and French press respectively. These findings, combined with the limited options of optimization for both bead beating and French press lysis as well as restricted potential for scaling up of bead beating lead to the decision to focus our optimization efforts on sonication as lysis method.</p> | + | <p>Our initial test showed that cell lysis by sonication yields extract with up to 2.8- and 4.4 fold higher expression than cell lysis by bead beating and French press respectively. These findings combined with the limited options of optimization for both bead beating and French press lysis as well as restricted potential for scaling up of bead beating, lead to the decision to focus our optimization efforts on sonication as lysis method.</p> |
| | </div> | | </div> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| Line 221: |
Line 233: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>Cell lysis by sonication has two main parameters that can be varied: the amplitude, which correlates to the power emitted by the sonication device and cycle number, representing the total time of sonication. Based on Yong-Chan Kwon & Michael C. Jewett we decided to use 10 s pulses with 15 s pause between pulses to prevent excessive heating of the sample. | + | <p>Cell lysis by sonication has two main parameters that can be varied: the amplitude, which correlates to the power emitted by the sonication device and cycle number, representing the total time of sonication. Based on Yong-Chan Kwon & Michael C. Jewett<sup><a href="#phareferences">2</a></sup> we decided to use 10 s pulses with 15 s pause between pulses to prevent excessive heating of the sample. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 249: |
Line 261: |
| | <p> | | <p> |
| | <br> | | <br> |
| − | Our screening revealed that increase of amplitude as well as increase of cycle number leads to higher protein content. The TXTL test showed high fluorescence intensity only for settings with low cycle numbers. This indicates that prolonged sonication damages the cellular machinery, declining the ability for protein expression. | + | Our screening revealed that increase in amplitude as well as increase in cycle number leads to higher protein content. The TXTL test showed high fluorescence intensity only for settings with low cycle numbers. This indicates that prolonged sonication damages the cellular machinery, reducing its ability for protein expression. |
| | </p> | | </p> |
| | <p> | | <p> |
| − | The highest-performing extracts where those obtained by sonication at 4 cycles at 33 % amplitude and 5 cycles at 10 % amplitude. Therefore we focused our next steps on further optimization of those settings. | + | The highest-performing extracts were those obtained by sonication with 4 cycles at 33 % amplitude and 5 cycles at 10 % amplitude. Therefore, we focused our next steps on further optimization of those settings. |
| | </p> | | </p> |
| | | | |
| Line 258: |
Line 270: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>In the last step we made an effort to scale up the sonication step. Preliminary experiments (results not shown) indicated 4 mL as the maximal sample volume that could efficiently be lysed with the device we had at our disposal. As indicated in the publication by Yong-Chan Kwon & Michael C. Jewett2 the number of sonication cycles was increased in proportion to the increase of sample volume. The presented results shown in the figure below shows that this approach was successful: Using higher sample volumes for sonication increased protein content and expression quality of the cell extract. This proves that upscaling of cell extract preparation with cell lysis by sonication is indeed feasible. | + | <p>In the last step we made an effort to scale up the sonication step. Preliminary experiments (results not shown) indicated 4 mL as the maximal sample volume that could be lysed efficiently with the device we had at our disposal. As indicated in the publication by Yong-Chan Kwon & Michael C. Jewett the number of sonication cycles was increased in proportion to the increase of sample volume. The presented results shown in the figure below show that this approach was successful: Using higher sample volumes for sonication increased protein content and expression quality of the cell extract. This proves that upscaling of cell extract preparation with cell lysis by sonication is indeed feasible. |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 286: |
Line 298: |
| | <p> | | <p> |
| | <br> | | <br> |
| − | Even though previous attempts by Shresta et al.[ref 3] to use Lysozyme in cell extract preparation have been unsuccessful, we tested the effect of adding the cell-wall degrading enzyme at 1 mM concentration to the cell lysis reaction. In our experiments lysozyme increases the efficiency of cell lysis for all tested settings. Moreover, the expression quality was likewise increased after Lysozyme addition, indicating that the remaining enzyme does not interfere with protein biosynthesis. | + | Even though previous attempts by Shresta et al.<sup><a href="#phareferences">3</a></sup> to use Lysozyme in cell extract preparation have been unsuccessful, we tested the effect of adding the cell-wall degrading enzyme at 1 mM concentration to the cell lysis reaction. Across all our experiments, lysozyme increased the efficiency of cell lysis for all tested settings. Moreover, the expression quality was likewise increased after Lysozyme addition, indicating that the remaining enzyme does not interfere with protein biosynthesis. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 295: |
Line 307: |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | Cultivation and cell lysis are the apparent most crucial steps in preparation of cell extract but there are other steps that can have a great impact onto cell extract quality. Some of our findings regarding those steps are: | + | Cultivation and cell lysis are the apparent most crucial steps in preparation of cell extract but there are other steps that can have a great impact on cell extract quality. Some of our findings regarding those steps are: |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 303: |
Line 315: |
| | <ul style=""> | | <ul style=""> |
| | <li>Freezing cell pellets overnight for next-day processing does not interfere with extract quality but confers a major simplification of cell extract preparation</li> | | <li>Freezing cell pellets overnight for next-day processing does not interfere with extract quality but confers a major simplification of cell extract preparation</li> |
| − | <li>Preliminary results indicate that omitting the run-off reaction after lysis does not significantly impair the extract quality</li> | + | <li>Preliminary results indicate that omitting the run-off reaction after lysis does not significantly impair extract quality</li> |
| − | <li>Dialysis of cell extract can be substituted by diafiltration in centrifugal filters without reducing expression yield of the resulting extract </li> | + | <li>Dialysis of cell extract can be substituted by diafiltration in centrifugal filters without reducing the expression yield of the resulting extract </li> |
| | </ul> | | </ul> |
| | </div> | | </div> |
| Line 311: |
Line 323: |
| | <hr> | | <hr> |
| | | | |
| | + | <h3>Characterizing Cell Extract Quality </h3> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | In order to validate the success of our optimization efforts, adequate characterization of cell extract is indispensable. |
| | + | Here we will give a short review about the tools for quality assessment we implemented. Further details about characterization |
| | + | of these tools are available on the <a href="https://2018.igem.org/Team:Munich/Measurement">Measurement</a> page. |
| | + | </p> |
| | + | <p> |
| | + | We had initially defined increase of protein content in cell extract as optimization goal. However, as we realized early on, protein content alone is not a good measure for extract quality, because it provides no information about the activity of the cellular machinery. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p> |
| | + | To assess functionality of cell extract, testing of protein expression is necessary, which can be achieved by expressing fluorescent proteins and measuring the fluorescence time trace in a plate reader. To find the optimal quality control we compared the performance of different fluorescent proteins in cell extract. |
| | + | </p> |
| | + | <p> |
| | + | We chose expression of mTurquoise – a variant of Cyan Fluorescent Protein (CFP) – as our favorite quality control for cell extract, as it results in high fluorescence intensity in a reproducible manner. |
| | + | </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/e/e0/T--Munich--Results-WL2_Four_Fluorescence.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p> |
| | + | In addition, to examining protein expression we decided to analyze transcription uncoupled from translation by transcribing an RNA aptamer that binds a fluorescent dye. |
| | + | <p> |
| | + | We compared the fluorescence time trace of a Malachite Green binding aptamer in different cell extract samples. The fluorescence time traces decline after 30-50min, indicating that RNA degradation starts to prevail over transcription. Differences in the observed kinetics can be explained by variations in cell extract composition. |
| | + | </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/e/e3/T--Munich--WL2_MG_aptamer.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | | | |
| | <h3> | | <h3> |
| Line 320: |
Line 377: |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| | <p> | | <p> |
| − | The main question was, if the different performance of the cell extracts (P15, E10, myTXTL), especially concerning phage titers, is due to a divergent composition or caused by a discrepancy in activity of the relevant proteins e.g. transcription and translation related proteins | + | The main question was if the different performance of the cell extracts (P15, E10, myTXTL), especially concerning phage titers, is due to a divergent composition or caused by a discrepancy in activity of the relevant proteins e.g. transcription and translation related proteins |
| | Therefore, one student was sent to Bavarian Biomolecular Mass Spectrometry Centre (BayBioMS), where he analyzed the samples under the supervision of Dr. Christina Ludwig. | | Therefore, one student was sent to Bavarian Biomolecular Mass Spectrometry Centre (BayBioMS), where he analyzed the samples under the supervision of Dr. Christina Ludwig. |
| | </p> | | </p> |
| − | <p>The results of the Mass Spectrometry gave us insight into the composition of the different TX-TLs. We were able to identify 1771 proteins in all the extracts. The results indicate a high homogeneity between all three TX-TLs, as illustrated in the heatmap. | + | <p>The results of the Mass Spectrometry gave us insight into the composition of the different TXTLs. We were able to identify 1771 proteins in all the extracts. The results indicate a high homogeneity between all three TXTLs, as illustrated in the heatmap. |
| − | Based on the LFQ values (Label Free Quantification) a volcano plot of two samples (Arbor/E10, Arbor/P15 and E10/P15) was generated. In general, there is no protein more frequent in Arbor TX-TL in comparison to E10 and P15. In contrast, some translation related proteins were slightly more abundant in E10 and P15 like certain tRNA ligases (PheS and PheT). The RecBCD subunits (the dsDNA degrading complex) is of similar abundance in all TX-TLs, showing the importance to consider its negative impact on the assembly. The results were further validated with DAVID, a tool for finding metabolic pathways based on proteomic data. The identified pathways indicate no correlation with transcription/translation related pathways (data not shown). | + | Based on the LFQ values (Label Free Quantification) a volcano plot of two samples (Arbor/E10, Arbor/P15 and E10/P15) was generated. In general, there is no protein more frequent in Arbor TXTL in comparison to E10 and P15. In contrast, some translation related proteins were slightly more abundant in E10 and P15 like certain tRNA ligases (PheS and PheT). The RecBCD subunits (the dsDNA degrading complex) is of similar abundance in all TXTLs, showing the importance to consider its negative impact on the assembly. The results were further validated with DAVID, a tool for finding metabolic pathways based on proteomic data. The identified pathways indicate no correlation with transcription/translation related pathways (data not shown). |
| | </p> | | </p> |
| | <p> | | <p> |
| − | We concluded that the performance difference is most likely due to decreased activity during TX-TL preparation rather than a change in composition. | + | We concluded that the performance difference is most likely due to decreased activity during TXTL preparation rather than a change in composition. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 359: |
Line 416: |
| | </div> | | </div> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <figcaption class="figure-caption">Volcano plots of myTXTL vs. P15 (A), myTX-TL vs. E10 (B) and E10 vs. P15 (C), LC-MS/MS measurements of the E. coli proteome lysates were performed in duplicates on a Q-Exactive HFX instrument in data-dependent acquisition mode (Top18 method). Total protein injection amount was 0.5 µg. Gradient length was 1h. The data was then analyzed using MaxQuant and the Uniprot E. coli K12 fasta file. The LFQ data was filtered for proteins that occur in both duplicates of at least one cell lysate group (Arbor E10 P14).</figcaption> | + | <figcaption class="figure-caption">Volcano plots of myTXTL vs. P15 (A), myTXTL vs. E10 (B) and E10 vs. P15 (C), LC-MS/MS measurements of the E. coli proteome lysates were performed in duplicates on a Q-Exactive HFX instrument in data-dependent acquisition mode (Top18 method). Total protein injection amount was 0.5 µg. Gradient length was 1h. The data was then analyzed using MaxQuant and the Uniprot E. coli K12 FASTA file. The LFQ data was filtered for proteins that occur in both duplicates of at least one cell lysate group (Arbor E10 P14).</figcaption> |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 371: |
Line 428: |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | With regard to global application of Phactory it is crucial to enable long-term storage and shipping of our cell extract. In order to fulfill these requirements we chose to freeze-dry our cell extract. Lyophilized extract can be stored at ambient temperature and can be reactivated by addition of only nuclease-free water. | + | With regard to global application of Phactory it is crucial to enable long-term storage and shipping of our cell extract. In order to fulfill these requirements, we chose to freeze-dry our cell extract. Lyophilized extract can be stored at ambient temperature and can be reactivated by addition of only nuclease-free water. |
| | | | |
| | Initial tests for the optimal lyophilization conditions revealed several important points: | | Initial tests for the optimal lyophilization conditions revealed several important points: |
| Line 383: |
Line 440: |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| Line 401: |
Line 457: |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| − |
| |
| | | | |
| | <h2> | | <h2> |
| Line 408: |
Line 462: |
| | </h2> | | </h2> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| | Phages were manufactured based on three components: the cell extract, an energy solution (ATP, GTP, NAD+ etc.) and a supplement | | Phages were manufactured based on three components: the cell extract, an energy solution (ATP, GTP, NAD+ etc.) and a supplement |
| − | solution (aminoacids, tRNAs, pholic acid etc.). The only additional component for phage assembly is pure phage DNA. We expanded the assembly platform by | + | solution (amino acids, tRNAs, pholic acid etc.). The only additional component for phage assembly is pure phage DNA. We expanded the assembly platform by |
| | </p> | | </p> |
| | </div> | | </div> |
| | </div> | | </div> |
| | | | |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ul style=""> | | <ul style=""> |
| Line 428: |
Line 482: |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | <h3> |
| | + | DNA Purification |
| | + | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p> An essential part of bacteriophage manufacturing is the DNA. We extracted it by Phenol-Chloroform precipitation. The Purification was controlled with an agarose gel, where two distinct bands (the proper high-mass band and a low-mass band) were present for all four samples. |
| | + | </p> |
| | + | <p> To reduce the amount of the low-mass bands, the protocol was changed to a column-based technique. With this technique the incorrect band was significantly reduced in all samples, which was proven with an agarose gel.</p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/f/fe/T--Munich--agarose_gel_dirty_phage.jpeg " class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> The agarose gel shows the DNA of a phenol chloroform purification of the bacteriophage genomes from left to right T7, GEC-3S, T4 and NES. The ladder in the first lane from left is the 1kb extended range ladder from NEB.</figcaption> |
| | + | </figure> |
| | + | |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/7/77/T--Munich--agarose_gel_clean_phage.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> The agarose gel shows the DNA of bacteriophages which was column purified. The bands are from left to right, a the 1kb extended range ladder from NEB the CLB-P2, CLP-P3, T4, T5, T7, NES, FFP and GEC-3S bacteriophage. |
| | + | </figcaption> |
| | + | </figure> |
| | + | |
| | + | |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | | | |
| | <h3> | | <h3> |
| Line 439: |
Line 521: |
| | </p> | | </p> |
| | <p> | | <p> |
| − | All in all our extract has the same phage assembly potential as the commercial cell extract. Moreover, lyophylization, which makes | + | All in all, our extract has the same phage assembly potential as the commercial cell extract. Moreover, lyophilization, which makes |
| − | the cell extract more long lived, does not reduce the effiency of bacteriophage assembly. We concluded that phages could be assembled on site in a durable lyophilized cell extract. | + | the cell extract better storable, does not reduce the efficiency of bacteriophage assembly. We concluded that phages could be assembled on site in a durable lyophilized cell extract. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 461: |
Line 543: |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | Phactory has is the ability to assemble various bacteriophages, in a bacteria-independent manner. To underline this feature and demonstrate universal applicability, we assembled a variety of different E. coli phages, both DNA and RNA-based. | + | Phactory has the ability to assemble various bacteriophages, in a bacteria-independent manner. To underline this feature and demonstrate universal applicability, we assembled a variety of different E. coli phages, both DNA and RNA-based. |
| | </p> | | </p> |
| | <br> | | <br> |
| Line 474: |
Line 556: |
| | | | |
| | <p> | | <p> |
| − | Pathogenic bacteria such as salmonella, pseudomonas and staphylococcus are prone to develop multi-drug resistance and pose an urgent or serious threat (Centers for Disease Control and Prevention, 2013. Antibiotic/Antimicrobial Resistance.). Therefore, to fulfill this medical need, phages specific for these bacteria should be assembled next in our cell-free system. Potentially, this would require co-expression of the respective sigma factors that are needed for initiation of transcription. | + | Pathogenic bacteria such as salmonella, pseudomonas and staphylococcus are prone to develop multi-drug resistance and pose an urgent or serious threat (Centers for Disease Control and Prevention, 2013. Antibiotic/Antimicrobial Resistance.). Therefore, to fulfill this medical need, phages specific for these bacteria should be assembled next in our cell-free system. |
| | </p> | | </p> |
| | | | |
| Line 533: |
Line 615: |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | + | <div class="col-12 col-md-3"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/54/T--Munich--GEC_3S_movie.gif" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure." rotate="90"> |
| | + | <figcaption class="figure-caption"> 3D reconstruction of Transmission electron microscopy images of the GEC-3S bacteriophage. </figcaption> |
| | + | </figure> |
| | + | </div> |
| | </div> | | </div> |
| | <div class="row"> | | <div class="row"> |
| Line 547: |
Line 635: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>To prove the influence of the DNA concentration on the bacteriophage titer, cell extract reactions were prepared with varying T4 DNA concentrations. The bacteriophage production was performed. The titer of the bacteriophages was measured with the top agar method and the formed plaques were counted. The increase in DNA concentration results also in an increase in the bacteriophage concentration. This increase is nonlinear and our model predicted. This finding is probably due to a critical concentration of phage proteins, which have to be reached for capsid head assembly (similar to a critical micelle concentration). | + | <p>To prove the influence of the DNA concentration on the bacteriophage titer, cell extract reactions were prepared with varying T4 DNA concentrations. The bacteriophage production was performed. The titer of the bacteriophages was measured with the top agar method and the formed plaques were counted. The increase in DNA concentration results in an increase in the bacteriophage concentration. This increase is nonlinear as our <a href="https://2018.igem.org/Team:Munich/Model">model</a> predicted. This finding is probably due to a critical concentration of phage proteins, which have to be reached for capsid head assembly (similar to a critical micelle concentration). |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 572: |
Line 660: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>The DNA sequence added to the cell-free system serves as the template for the required phage. We saw, that DNA-Polymerases can amplify the DNA segment, multiplying the amount of DNA in the cell-free reaction. | + | <p>The DNA sequence added to the cell-free system serves as the template for the required phage. We saw, that DNA-Polymerases can amplify the DNA segment, multiplying the amount of DNA in the cell-free reaction.<sup><a href="#phareferences">4</a></sup> |
| | </p> | | </p> |
| | <p> | | <p> |
| | To assess this effect and its dependence on deoxynucleotide triphosphates (dNTPs), we performed an absolute quantification of T7 DNA in the cell-free reaction by quantitative PCR (qPCR). A standard curve with a serial dilution of T7 DNA. We used the TXTL qPCR protocol (add link). | | To assess this effect and its dependence on deoxynucleotide triphosphates (dNTPs), we performed an absolute quantification of T7 DNA in the cell-free reaction by quantitative PCR (qPCR). A standard curve with a serial dilution of T7 DNA. We used the TXTL qPCR protocol (add link). |
| − | We used two selected cell extracts from us (P15 and E10), which reached similar phage titers compared to the commercial cell extract (myTXTL) in previous experiments. | + | We used two selected cell extracts (P15 and E10), which reached similar phage titers compared to the commercial cell extract (myTXTL) in previous experiments. |
| | We compared the replication potential in comparison to myTXTL. | | We compared the replication potential in comparison to myTXTL. |
| | </p> | | </p> |
| | <p> | | <p> |
| − | The addition of dNTP to the reference reaction leads to an increase in DNA concentration by a factor of 15 in the reaction after 4 hours (290 ng compared to 19 ng). This is higher than in the myTXTL reaction without additional dNTPs, in which there is a 1.8-fold increase in DNA (91 ng compared to 51 ng) after the 4-hour reaction. | + | The addition of dNTP to the reference reaction leads to an increase in DNA concentration by a factor of 15 in the reaction after 4 hours (290 ng compared to 19 ng). This is higher than in the myTXTL reaction without additional dNTPs, where a 1.8-fold increase in DNA (91 ng compared to 51 ng) after the 4-hour reaction is observed. |
| | The home-made cell extracts P10 and E15 however do not resemble this behavior. | | The home-made cell extracts P10 and E15 however do not resemble this behavior. |
| | </p> | | </p> |
| Line 591: |
Line 679: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/3/36/T--Munich--Results_WL2_Qpcr.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/3/36/T--Munich--Results_WL2_Qpcr.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">DNA replication within cell extracts, E15 and P15 are self prepared cell extracts, which reached similar phage titers compared to the commercial cell extract (myTXTL). ctrl = control without dNTPs. </figcaption> | + | <figcaption class="figure-caption">DNA replication within cell extracts, E15 and P15 are self-prepared cell extracts, which reached similar phage titers compared to the commercial cell extract (myTXTL). ctrl = control without dNTPs. </figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| − | <h3>Modelling Phage Production in Cell Extract</h3>
| |
| | | | |
| | <h2> | | <h2> |
| | Modular Bacteriophage Composition | | Modular Bacteriophage Composition |
| | </h2> | | </h2> |
| − | | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| − | <div class="row"> | + | |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>
| + | <p>Bacteriophage modularity can be achieved by protein engineering without changing the genomic DNA of the bacteriophage and synthetic genome engineering. |
| − | In the TX-TL system should be possible to modify phage proteins without altering their genome. This was attempted by modifying HOC (highly immunogenic capsid protein), which is part of the capsid protein structure of the T4-phage. Therefore, His-TEV-YFP-HOC was separately expressed and the purified protein was applied to our phage assembly.
| + | |
| | </p> | | </p> |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | <h3> |
| | + | Protein engineering |
| | + | </h3> |
| | | | |
| − | <div class="row">
| + | |
| − | <div class="col-12"> | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/d/d8/T--Munich--Results-His_YFP_HOC_front.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">His-YFP-HOC protein structure.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | <p> | | <p> |
| − | For protein modification of the T4 capsid protein HOC, a plasmid for protein expression was cloned and His-YFP-HOC (82kDa) was expressed. The plasmid was transformed into BL21, expressed and puryfied by nickel affinity chromatography and gelfiltration. | + | In the TXTL system, it should be possible to modify phage proteins without altering their genome. This was attempted by modifying HOC (highly immunogenic capsid protein), which is part of the capsid protein structure of the T4-phage. Therefore, His-TEV-YFP-HOC was separately expressed and the purified protein was applied to our phage assembly.<sup><a href="#phareferences">5</a></sup> |
| − | </p>
| + | </p> |
| − | <p>FILM</p>
| + | <p> |
| − | <p>MODEL</p>
| + | For protein modification of the T4 capsid protein HOC, a plasmid for protein expression was cloned and His-YFP-HOC (82kDa) was expressed. The plasmid was transformed into BL21, expressed and purified by nickel affinity chromatography and gel filtration. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | + | |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 627: |
Line 725: |
| | <h5 class="mb-0"> | | <h5 class="mb-0"> |
| | <button class="btn btn-link collapsed" data-toggle="collapse" data-target="#collapseOne" aria-expanded="false" aria-controls="collapseOne"> | | <button class="btn btn-link collapsed" data-toggle="collapse" data-target="#collapseOne" aria-expanded="false" aria-controls="collapseOne"> |
| − | <span class="pl-3" style="color: white;">Protein Purificaton</span> | + | <span class="pl-3" style="color: white;">Protein Purification</span> |
| | </button> | | </button> |
| | </h5> | | </h5> |
| Line 706: |
Line 804: |
| | </figure> | | </figure> |
| | </div> | | </div> |
| − | </div> | + | </div> |
| − | | + | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| | </div> | | </div> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <figcaption class="figure-caption">Bacteria infected with (A) and without (B) YFP modified phages </figcaption> | + | <figcaption class="figure-caption">Bacteria infected without (left) and with (right) YFP modified phages </figcaption> |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | |
| | + | <h3> |
| | + | Purification of the modified T4 bacteriophage |
| | + | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>To show the possibility of our manufacturing system we modified our T4 bacteriophage at the Hoc protein with the fusion of a His-YFP. In this experiment the His-tag was utilized to purify the bacteriophage solution from the cell extract after the assembly process. </p> |
| | | | |
| | + | <p>The bacteriophages with the His-tag, were loaded on Ni-NTA beads which where embedded in chromatography columns. After four subsequently washing steps imidazole was used to elute the His-tag from the beads. As a negative control, bacteriophages without a His-tag were treated equally. </p> |
| | + | |
| | + | |
| | + | <p>To test the purification a spot assay was performed. On the left there are the T4 bacteriophages which were eluted from the sample with the His-Tagged bacteriophages and on the right side there is negative control with the native eluted bacteriophages. The titer of the bacteriophages is lower in the right sample, due to the fact that single plaques are visible, which indicates a low concentration. In contrast the His-tagged bacteriophages, which have a higher concentration, were only one large lysis area is visible. </p> |
| | | | |
| − | <div class="row">
| |
| − | <div class="col-12">
| |
| − | <p>
| |
| − | The purified protein was added to the assembly mix. Bacteria were transfected with the modified phages. Unbound phages and protein were removed by centrifugation. Fluorescence was measured in dependence of the proximity to the bacteria. Theoretically, YFP intensity should correlate with the binding of YFP-HOC modified phages to the bacteria.
| |
| − | </p>
| |
| | </div> | | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/1/10/T--Munich--HisTag_spottest.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption">Spot test of the engineered bacteriophages after His-tag purification, the left spot is the T4 phage, which is modified at the Hoc-Protein and the right spot is the negative control, where native T4 phages were used.</figcaption> |
| | + | </figure> |
| | </div> | | </div> |
| | + | </div> |
| | + | |
| | + | |
| | + | |
| | | | |
| | | | |
| | <hr> | | <hr> |
| | + | |
| | + | <h3> |
| | + | Phage Genome Engineering |
| | + | </h3> |
| | + | |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>An additional advantage of Phactory is the possibility of rebooting bacteriophages from their genomic template, which is especially important for genetic engineering of bacteriophages. With home-made cell extract it was possible to manufacture an engineered MS2 RNA phage, where a polyhistidine-tag was added on the phage RNA polymerase. The genomic template was generated via gibson assembly and PCR amplification. After purification of the engineered genome, phages were assembled in our self-made P15 cell extract. A plaque assay confirmed the successful assembly of functional phages with a titer of 3 × 10<sup>7</sup> PFU/ml.</p> |
| | + | <p>To test the his-tag modification, the Phages were amplified in a bacterial culture flask. After lysis of the bacteria the tagged polymerase was purified from the supernatant by nickel affinity chromatography. An SDS-PAGE proved that the 62 kDa Protein remained in the Nickel column due to the successfully engineering of an inserted His-tag. </p> |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | <figure class="figure"> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/52/T--Munich--MS2_gel_enge.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure." height=”50%” width="50%" > |
| | + | <figcaption class="figure-caption">SDS gel of the His-tag purified MS2 RNA dependent RNA polymerase</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | | | |
| | | | |
| Line 732: |
Line 862: |
| | Quality Control | | Quality Control |
| | </h2> | | </h2> |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| Line 739: |
Line 869: |
| | </div> | | </div> |
| | </div> | | </div> |
| − | <div class="row"> | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ul style=""> | | <ul style=""> |
| Line 757: |
Line 887: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>We performed a Plaque Assay to determine the titer of viable phages in our assembly batch. By creating serial dilutions, we were able to calculate a plaque forming units/milliliter (PFU/ml) value. The plaque assay protocol (link) was used. | + | <p>We performed a Plaque Assay to determine the activity of the viable phages (titer) in our assembly batch. By creating serial dilutions, we were able to calculate a plaque forming units/milliliter (PFU/ml) value. The <a href="https://static.igem.org/mediawiki/2018/2/23/T--Munich--AgarOverlayPlaqueAssay_.pdf "> plaque assay protocol </a> was used. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 767: |
Line 897: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/d/d0/T--Munich--Results_Wl3_T7_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/d/d0/T--Munich--Results_Wl3_T7_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption"> </figcaption> |
| | + | |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 775: |
Line 906: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/b/b5/T--Munich--Results_Wl3_MS2_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/b/b5/T--Munich--Results_Wl3_MS2_plaque.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption"></figcaption> | + | <figcaption class="figure-caption"> Plaque Assay of manufactured T7 DNA -bacteriophage (top) and MS2 RNA-bacteriophage (bottom) in our self-produced cell extract P15</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 806: |
Line 937: |
| | <div class="col-12"> | | <div class="col-12"> |
| | <p> | | <p> |
| − | To rapidly detect functionality of the phages, Reverse Transcription-quantitative PCR (RT-qPCR) was applied to pellets of T7-infected host cells at different timepoints. The RT-qPCR protocol was used (add link). Using the delta-delta-Ct method3, relative expression was determined and normalized to the value at 3 minutes after addition of the phages. Whereas expression of E. coli genes remains stable, expression of three T7 genes is elevated throughout the experiment. The change of expression of all three phage genes reaches a first peak after 12 minutes and a second peak after 18 or 21 minutes. | + | To rapidly detect functionality of the phages, reverse transcription-quantitative PCR (RT-qPCR) was applied to pellets of T7-infected host cells at different timepoints. The RT-qPCR protocol was used. Using the delta-delta-Ct method3, relative expression was determined and normalized to the value at 3 minutes after addition of the phages. Whereas expression of E. coli genes remains stable, expression of three T7 genes is elevated throughout the experiment. The change of expression of all three phage genes reaches a first peak after 12 minutes and a second peak after 18 or 21 minutes. |
| | </p> | | </p> |
| | <p> | | <p> |
| − | The sharp increase of phage gene expression in the first phase of the experiment displays the ability of the phages to successfully infect the bacteria and initiate reproduction. The second and even higher increase of expression is likely attributed to a second wave of infection of already replicated phages. This indicates that the phages are capable of reproducing inside their host bacteria, resulting in multiplication of functional phages. | + | The sharp increase of phage gene expression in the first phase of the experiment displays the ability of the phages to successfully infect the bacteria and initiate reproduction. The second and even higher increase of expression is likely attributed to a second wave of infection of previously replicated phages. This indicates that the phages are capable of reproducing inside their host bacteria, resulting in multiplication of functional phages. |
| | The increase of expression of T7P01 is more pronounced than that of T7P07, which is in turn is stronger than that of T7P29. This circumstance is likely caused by differences in primer efficacy, a value describing the doubling rate in between every PCR cycle. For better reliability of the RT-qPCR quality control, primer efficacy should be assessed by creating a standard curve. | | The increase of expression of T7P01 is more pronounced than that of T7P07, which is in turn is stronger than that of T7P29. This circumstance is likely caused by differences in primer efficacy, a value describing the doubling rate in between every PCR cycle. For better reliability of the RT-qPCR quality control, primer efficacy should be assessed by creating a standard curve. |
| | </p> | | </p> |
| Line 822: |
Line 953: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Endotoxins are pyrogens deriving from gram-negative bacteria. Their mini from any pharmaceutical product is mandatory. Therefore, or Phactory, we engineered an E. coli strain lacking lipid A, a major endotoxin component and used this bacterium to produce our cell extract To evaluate endotoxin content of different cell extracts, a Limulus Amebocyte Lysate (LAL)-test was performed according to the <a href="https://www.genscript.com/product/documents?cat_no=L00350&catalogtype=Document-PROTOCOL">supplier manual</a>. As a reference, we compared the cell extract from our msbB-deficient strain (K2) to a cell extract from a wild-type strain (K4) as well as a commercial cell-free system (myTXTL, Arbor Biosciences). A solution with live <i>E. coli</i> served as a positive control.</p> | + | <p>Endotoxins are pyrogens deriving from gram-negative bacteria. Their removal from any pharmaceutical product is mandatory. Therefore, we engineered an E. coli strain lacking lipid A, a major endotoxin component and used this bacterium to produce our cell extract. To evaluate endotoxin content of different cell extracts, a Limulus Amebocyte Lysate (LAL)-test was performed according to the <a href="https://www.genscript.com/product/documents?cat_no=L00350&catalogtype=Document-PROTOCOL">supplier manual</a>. As a reference, we compared the cell extract from our msbB-deficient strain (K2) to a cell extract from a wild-type strain (K4) as well as a commercial cell-free system (myTXTL, Arbor Biosciences). A solution with lysed <i>E. coli</i> served as a positive control.</p> |
| | <p>Compared to the K4 strain our msbB-deficient K2 cell extract had 49-fold reduced endotoxin levels (0.06 EU/ml compared to 2.94 EU/ml). Other cell extracts such as the P15 cell extract (3.83 EU/ml) and the commercial myTXTL (4.65 EU/ml) had even higher endotoxin contents.</p> | | <p>Compared to the K4 strain our msbB-deficient K2 cell extract had 49-fold reduced endotoxin levels (0.06 EU/ml compared to 2.94 EU/ml). Other cell extracts such as the P15 cell extract (3.83 EU/ml) and the commercial myTXTL (4.65 EU/ml) had even higher endotoxin contents.</p> |
| | </div> | | </div> |
| Line 841: |
Line 972: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| − | <p>Removal of endotoxins is impeded by their tendency to form stable interactions with other biomolecules2. Our method of preventing the lipid A biosynthesis is therefore superior to extensive isolation steps required for removing endotoxins in conventional phage production. </p> | + | <p>Removal of endotoxins is impeded by their tendency to form stable interactions with other biomolecules. Our method of preventing the lipid A biosynthesis is therefore superior to extensive isolation steps required for removing endotoxins in conventional phage production. </p> |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | <h3> |
| | + | Nanopore Sequencing Enhances DNA Purification |
| | + | </h3> |
| | + | <div class="row"> |
| | + | <div class="col-12 col-md-6"> |
| | + | <p>Besides the cell extract, the DNA quality is the key to phage assembly. Impure DNA (Host DNA, proteins contamination) complicates the assembly of the phage within the cell lysate. A highly sensitive method to measure DNA is Oxford Nanopore sequencing.<sup><a href="#phareferences">6</a></sup> |
| | + | </p> |
| | + | |
| | + | <p>Our first attempts with classical chloroform/phenol extraction failed because of low molecular DNA (agarose gel). Therefore, we decided to search for other purification protocols, DNAse I and the Norgene kit (46800) tremendously increased DNA purity. With this DNA, we were able to achieve high titers by assembly. |
| | + | </p> |
| | + | |
| | + | </div> |
| | + | <div class="col-12 col-md-6"> |
| | + | |
| | + | <figure class="figure"> |
| | + | <img src=" https://static.igem.org/mediawiki/2018/1/16/T--Munich--contamination_file_seq.jpeg" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| | + | <figcaption class="figure-caption"> Illustration of DNA composition after purification, 3S phage DNA was purified from the <i>E. coli</i> host by chloroform-phenol (CM) and column extraction with and without DNAse I treatment after cell lysis.</figcaption> |
| | + | </figure> |
| | + | </div> |
| | + | </div> |
| | + | |
| | + | |
| | | | |
| | <h3> | | <h3> |
| Line 891: |
Line 1,044: |
| | | | |
| | <p> | | <p> |
| − | The main task after receiving genomes was to assemble the phage genomes from the Nanopore reads without the reference | + | The main task after receiving genomes was to assemble the phage genomes from the Nanopore reads without the reference |
| − | genome and gain the origin sequence. There are several tools available, but for minION Nanopore recommended tools are | + | genome and gain the origin sequence. There are several tools available but for minION Nanopore recommended tools are |
| − | canu and miniasm, of which we finally used the latter. | + | "canu" and "miniasm" of which we used the latter. |
| | </p> | | </p> |
| | <p> | | <p> |
| − | Despite an excellent lab team we have been confronted with a major problem: contamination. Contamination is a problem | + | Despite an excellent lab team, we have been confronted with a major problem: contamination. Contamination is a problem |
| − | regarding the therapeutic usage of the final phage extract, but also an important factor in the bioinformatics pipeline | + | regarding the therapeutic usage of the bacteriophages, but also an important factor in any bioinformatics pipeline |
| − | because contamination-based reads can confuse the assembly process. We thus developed <a href="https://2018.igem.org/Team:Munich/Software">sequ-into</a> to detect | + | because contamination-based reads can confuse the assembly process. We thus developed <a href="https://2018.igem.org/Team:Munich/Software">Sequ-Into</a> to detect |
| − | the contamination and also get rid of contamination-originated reads. In the context of our project, several DNA | + | the contamination and also get rid of contamination-originated reads. In the context of our project, several DNA |
| − | purification protocols were evaluated with sequ-into and allowed iterative engineering cycles in Phactory, leading to | + | purification protocols were evaluated with Sequ-Into. This allowed iterative engineering cycles in Phactory, leading to |
| | unparalleled purity of up to 96% (bases sequenced). | | unparalleled purity of up to 96% (bases sequenced). |
| | </p> | | </p> |
| | <p> | | <p> |
| − | While evaluating the sequencing data with sequ-into we noticed that in the first 10% of the sequencing time of | + | While evaluating the sequencing data with Sequ-Into, we noticed that in the first 10% of the sequencing time of |
| | each experiment, more non-target reads were sequenced than in any latter time interval. | | each experiment, more non-target reads were sequenced than in any latter time interval. |
| − | Moreover we found that this also holds true for the first x sequenced reads, which allowed us to setup sequ-into such | + | Moreover, we found that this also holds true for the first x sequenced reads, which allowed us to setup sequ-into such |
| − | that it only analyses the first 1000 reads of a minION sequencing experiment, speeding up computation and enabling an | + | that it only analyses the first 1000 reads of a minION sequencing experiment, speeding up computation and enabling |
| | en-run analysis. | | en-run analysis. |
| | </p> | | </p> |
| Line 921: |
Line 1,074: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/4/44/T--munich--rank_timeline.png" class="figure-img img-fluid rounded"> | | <img src="https://static.igem.org/mediawiki/2018/4/44/T--munich--rank_timeline.png" class="figure-img img-fluid rounded"> |
| − | <figcaption class="figure-caption">Also in the first x sequenced reads.</figcaption> | + | <figcaption class="figure-caption">Equal Results are shown for the first x sequenced reads.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 927: |
Line 1,080: |
| | | | |
| | <p> | | <p> |
| − | After getting rid of the contamination we noticed a non-uniform coverage in the phage genome assemblies after | + | After getting rid of the contamination, we noticed a non-uniform coverage in the phage genome assemblies after |
| | re-aligning the reads to the assembly. | | re-aligning the reads to the assembly. |
| | </p><p> | | </p><p> |
| − | Rearranging the middle part which initially was “over-expressed” and re-assembling led us to the expected uniform | + | Rearranging the middle part which initially was “over-expressed” and re-assembling led us to the expected uniform |
| | coverage of the de-novo genome sequence after re-aligning the input reads. | | coverage of the de-novo genome sequence after re-aligning the input reads. |
| | Finally, we could predict coding-sequences and visualize all descriptive statistics in genome diagrams. | | Finally, we could predict coding-sequences and visualize all descriptive statistics in genome diagrams. |
| Line 960: |
Line 1,113: |
| | </div> | | </div> |
| | <div></div> | | <div></div> |
| − | <p>More details at <a href="https://2018.igem.org/Team:Munich/DataAnalysis">Data Analysis</a> page.</p> | + | <p>More details at <a href="https://2018.igem.org/Team:Munich/Measurement">Data Analysis</a> page.</p> |
| | | | |
| | | | |
| Line 967: |
Line 1,120: |
| | Encapsulation | | Encapsulation |
| | </h2> | | </h2> |
| | + | |
| | + | <div class="row" style="background-color: rgba(200,200,200,1);"> |
| | + | <div class="col-12"> |
| | + | <p> |
| | + | After successful bacteriophage production and toxicity evaluation the final step of the manufacturing pipeline is therapeutic application. |
| | + | By nature, bacteriophages possess a relatively short have-life when orally administered. Encapsulation in alginate negates this effect by protecting phages from gastric fluid. <sup><a href="#phareferences">7</a></sup> |
| | + | </p> |
| | + | <p> |
| | + | We constructed a simple <a href="https://2018.igem.org/Team:Munich/Hardware">device</a> that allows us to encapsulate our bacteriophages in alginate droplets. |
| | + | </p> |
| | + | </div> |
| | + | </div> |
| | | | |
| | <h3>Droplets are Monodisperse</h3> | | <h3>Droplets are Monodisperse</h3> |
| Line 973: |
Line 1,138: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/8/84/T--Munich--Results_brightfield_droplet2_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/8/84/T--Munich--Results_brightfield_droplet2_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">perfectly circular droplets</figcaption> | + | <figcaption class="figure-caption">Alginate composition of 1.8 % Alginate and 0.2 % low-viscosity Alginate delivers spherical Alginate microspheres.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 979: |
Line 1,144: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/9/9c/T--Munich--Results_Droplets_zstack-400_8bit.gif" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/9/9c/T--Munich--Results_Droplets_zstack-400_8bit.gif" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">Animated Zoom Through Z-Stack</figcaption> | + | <figcaption class="figure-caption">Alginate encapsulated phages, DNA stained with SYBR Gold.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 985: |
Line 1,150: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/c/c8/T--Munich--Results_brightfield_droplet_disperse_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/c/c8/T--Munich--Results_brightfield_droplet_disperse_hardware.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">monodisperse droplets</figcaption> | + | <figcaption class="figure-caption">Monodisperse Alginate Droplets at 150X magnification.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 991: |
Line 1,156: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p> |
| | + | In order to achieve defined phage concentrations and therefore defined doses, we optimized the monodispersity of our alginate droplets. |
| | + | </p> |
| | + | <p> |
| | + | In our initial attempts to create alginate droplets the size within a batch often varied significantly. Additionally, a lot of droplets were lost due to |
| | + | aggregation. Optimization of parameters such as flow rate, |
| | + | alginate concentration and N2 pressure led to an increase of monodispersity for all tested sizes (50-300 μm). |
| | + | Specifically, an alginate concentration of 1.8 % alginate and 0.2 % low-viscosity alginate proved to be ideal. Pressure and flow rate determine the droplet sizes and can be varied. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 997: |
Line 1,169: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/3/38/T--Munich--Results_droplet_optimization.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/3/38/T--Munich--Results_droplet_optimization.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">Monodispersity of alginate droplets with three different diameters before and after optimization of the hardware device.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 1,009: |
Line 1,181: |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p>The main problem of oral application is the acidic environment in the gastric fluid, necessitating protective measures against degradation. The other requirement of phage protection is the release of functional phages in the intestine. For this reason, we compared the behavior of the encapsulated phages and non-encapsulated phages in simulated gastric fluid (SGF) and simulated intestinal fluid (SIF). |
| | + | </p> |
| | + | <p> |
| | + | In SGF, the number of active non-encapsulated phages decreases by more than 99.99 % within an hour. This shows the urgent need of a form of protection. As a reference, we used phages that were chemically released by citrate from alginate droplets. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 1,015: |
Line 1,190: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/4/4f/T--Munich--Results_Joe_SGF_Phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/4/4f/T--Munich--Results_Joe_SGF_Phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growth fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">Non-encapsulated phages are highly degraded within an hour in simulated gastric fluid.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| | </div> | | </div> |
| − |
| |
| − | <hr>
| |
| | | | |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12 col-md-6"> | | <div class="col-12 col-md-6"> |
| − | <p>Discussion goes here | + | <p> |
| | + | In comparison, the encapsulated phages were tested in SGF for the same time as the non-encapsulated phages. Afterwards, the same droplets were exposed for two hours to simulated intestinal fluid to test the release of functional bacteriophages in this environment. |
| | + | </p> |
| | + | <p>The encapsulated phages were hardly released in a timeframe of one hour when exposed to SGF. After transferring the capsules to SIF the number of active phages reached that of the reference. |
| | + | This indicates that the encapsulation of bacteriophages in alginate capsules enables the possibility of an oral application. Further experiments could test the alginate capsules in an animal model system. |
| | </p> | | </p> |
| | </div> | | </div> |
| Line 1,030: |
Line 1,207: |
| | <figure class="figure"> | | <figure class="figure"> |
| | <img src="https://static.igem.org/mediawiki/2018/a/a2/T--Munich--Results_Droplets_phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> | | <img src="https://static.igem.org/mediawiki/2018/a/a2/T--Munich--Results_Droplets_phages.png" class="figure-img img-fluid rounded" alt="A generic square placeholder image with rounded corners in a figure."> |
| − | <figcaption class="figure-caption">growh fermenter vs. shaking flask side by side with FI mtq2</figcaption> | + | <figcaption class="figure-caption">A small number of encapsulated phages are released in SGF. In SIF, functional phages are released in high concentration.</figcaption> |
| | </figure> | | </figure> |
| | </div> | | </div> |
| Line 1,036: |
Line 1,213: |
| | | | |
| | | | |
| − | | + | <!--- |
| | <div class="row"> | | <div class="row"> |
| | <div class="col-12"> | | <div class="col-12"> |
| Line 1,054: |
Line 1,231: |
| | </div> | | </div> |
| | </div> | | </div> |
| | + | ---> |
| | + | |
| | <hr> | | <hr> |
| | | | |
| | <div id="phareferences" class="row"> | | <div id="phareferences" class="row"> |
| − | <h2>References [FIX THIS!]</h2> | + | <h2>References</h2> |
| | <div class="col-12"> | | <div class="col-12"> |
| | <ol> | | <ol> |
| − | <li><a href="http://www.sciencedirect.com/science/article/pii/S0092867415012003?via%3Dihub">Zetsche, B, Gootenberg, J.S., Abudayyeh, O.O., Slaymaker, I.M., Makarova, K.S., Essletzbichler, P., Volz, S.E., van der Oost, J., Regev, Aviv, Koonin, E.V., Zhang, F., 2015. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759-771. </a></li> | + | <li><a href="https://www.jove.com/video/50762/protocols-for-implementing-an-escherichia-coli-based-tx-tl-cell-free"> Sun, Zachary Z., et al. "Protocols for implementing an Escherichia coli based TXTL cell-free expression system for synthetic biology." Journal of visualized experiments: JoVE 79 (2013). </a></li> |
| | + | <li><a href="https://www.nature.com/articles/srep08663"> Kwon, Yong-Chan, and Michael C. Jewett. "High-throughput preparation methods of crude extract for robust cell-free protein synthesis." Scientific reports 5 (2015): 8663. </a></li> |
| | + | <li><a href="https://www.future-science.com/doi/abs/10.2144/0000113924">Shrestha, Prashanta, Troy Michael Holland, and Bradley Charles Bundy. "Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing." Biotechniques 53.3 (2012): 163-174.</a></li> |
| | + | <li><a href="http://www.pnas.org/content/84/14/4767"> Modrich, P., and C. C. Richardson. "Bacteriophage T7 deoxyribonucleic acid replication invitro. Bacteriophage T7 DNA polymerase: an an emzyme composed of phage-and host-specific subunits." Journal of Biological Chemistry 250.14 (1975): 5515-5522. </a></li> |
| | + | <li><a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1563720/"> Sathaliyawala, Taheri, et al. "Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: a novel in vitro approach to construct multicomponent HIV vaccines." Journal of virology 80.15 (2006): 7688-7698. </a></li> |
| | + | <li><a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3804109/"> Maitra, Raj D., Jungsuk Kim, and William B. Dunbar. "Recent advances in nanopore sequencing." Electrophoresis 33.23 (2012): 3418-3428. </a></li> |
| | + | <li><a href="https://www.nature.com/articles/srep41441"> Colom, Joan, et al. "Microencapsulation with alginate/CaCO 3: A strategy for improved phage therapy." Scientific reports 7 (2017): 41441 </a></li> |
| | </ol> | | </ol> |
| | </div> | | </div> |