Cecilia 1403 (Talk | contribs) |
Cecilia 1403 (Talk | contribs) |
||
| Line 184: | Line 184: | ||
<figure style="text-align:center;"> | <figure style="text-align:center;"> | ||
<img src="https://static.igem.org/mediawiki/2018/2/27/T--Chalmers-Gothenburg--Project_Anchor.png" class="img-fluid" alt="anchoring representation"> | <img src="https://static.igem.org/mediawiki/2018/2/27/T--Chalmers-Gothenburg--Project_Anchor.png" class="img-fluid" alt="anchoring representation"> | ||
| − | |||
<p>In order to make it possible to locally treat the cancer, as well as to detect the tumor location, the yeast needs to bind specifically to the colon cancer cells. This is made possible through the use of Histone like protein A (HlpA) from the bacteria <i>Streptococcus gallolyticus</i>, which is commonly associated with colon cancer (Boleji et al. 2009; Ho et al., 2018). HlpA binds to the surface of the bacteria itself and also to heparan sulfate proteoglycans (HSPG), more specifically Syndecan 1 and possibly also Syndecan 2, on the surface of the colon cancer cell (Ho et al., 2018). In this way <i>S. gallolyticus</i> has been found to adhere to colon cancer cells and then infiltrate colon cancer tumors (Boleji et al. 2009). </p> | <p>In order to make it possible to locally treat the cancer, as well as to detect the tumor location, the yeast needs to bind specifically to the colon cancer cells. This is made possible through the use of Histone like protein A (HlpA) from the bacteria <i>Streptococcus gallolyticus</i>, which is commonly associated with colon cancer (Boleji et al. 2009; Ho et al., 2018). HlpA binds to the surface of the bacteria itself and also to heparan sulfate proteoglycans (HSPG), more specifically Syndecan 1 and possibly also Syndecan 2, on the surface of the colon cancer cell (Ho et al., 2018). In this way <i>S. gallolyticus</i> has been found to adhere to colon cancer cells and then infiltrate colon cancer tumors (Boleji et al. 2009). </p> | ||
| Line 202: | Line 201: | ||
<h1 style="text-align:center">Target molecule</h1> | <h1 style="text-align:center">Target molecule</h1> | ||
<a class="anchor" id="TargetM"></a> | <a class="anchor" id="TargetM"></a> | ||
| − | + | <figure style="text-align:center;"> | |
| + | <img src="https://static.igem.org/mediawiki/2018/thumb/e/ea/T--Chalmers-Gothenburg--Project_Pheromone.png/1600px-T--Chalmers-Gothenburg--Project_Pheromone.png" class="img-fluid"> | ||
| + | <br> | ||
<h1>Gas vesicles</h1> | <h1>Gas vesicles</h1> | ||
<a class="anchor" id="GasV"></a> | <a class="anchor" id="GasV"></a> | ||
<figure style="text-align:center;"> | <figure style="text-align:center;"> | ||
<img src="https://static.igem.org/mediawiki/2018/f/f3/T--Chalmers-Gothenburg--Project_GasVescicles.png" class="img-fluid alt="Gas vesicles representation" alt="anchoring representation"> | <img src="https://static.igem.org/mediawiki/2018/f/f3/T--Chalmers-Gothenburg--Project_GasVescicles.png" class="img-fluid alt="Gas vesicles representation" alt="anchoring representation"> | ||
| − | |||
<h2>Natural buoyancy</h2> | <h2>Natural buoyancy</h2> | ||
<p>Gas vesicles are hollow proteins complexes that take cylindrical shapes of 45 to 200 nm diameter (Walsby, 1994). In nature, gas vesicles are produced by a variety of aquatic microorganisms to control their buoyancy in water. A well-conserved operon of more than 10 genes is responsible for the production of these vesicles. The two most important genes are the genes Gas Vesicle Protein A or its homolog B and Gas Vesicle Protein C (GvpA/B and GvpC). They form the rigid structure of the gas vesicles (fig with GvpA and GvpC, check where it is from), which is permeable to gasses but not liquids (Sivertse et al, 2010). Thus, gas from the cytoplasm will diffuse into the vesicles while other components will be held out. The other genes present in the operon are less known but are thought to be mainly involved in regulation, folding and gas vesicle assembly. These proteins do not appear in the final structure (Sivertse et al, 2010).</p> | <p>Gas vesicles are hollow proteins complexes that take cylindrical shapes of 45 to 200 nm diameter (Walsby, 1994). In nature, gas vesicles are produced by a variety of aquatic microorganisms to control their buoyancy in water. A well-conserved operon of more than 10 genes is responsible for the production of these vesicles. The two most important genes are the genes Gas Vesicle Protein A or its homolog B and Gas Vesicle Protein C (GvpA/B and GvpC). They form the rigid structure of the gas vesicles (fig with GvpA and GvpC, check where it is from), which is permeable to gasses but not liquids (Sivertse et al, 2010). Thus, gas from the cytoplasm will diffuse into the vesicles while other components will be held out. The other genes present in the operon are less known but are thought to be mainly involved in regulation, folding and gas vesicle assembly. These proteins do not appear in the final structure (Sivertse et al, 2010).</p> | ||
Revision as of 21:11, 12 October 2018
Navigation
Project Overview
Our product is a pill containing a genetically engineered organism that can be used to detect and treat colon cancer. The organism should be able to localize cancer cells in the colon, locally kill the cells and report the presence of cancer. For this purpose, we have chosen to work with the yeast Saccharomyces boulardii and divided our project into three parts. The first part of the project concerns cancer cell localization, which is achieved with yeast to cancer cell anchoring. By making the yeast express Histone-like protein A on its surface it can specifically bind to cancer cells. In the second part of the project, we make the yeast secrete a target molecule with the purpose to treat cancer. Here, we exploit the native mating system of yeast in order to express the target molecule only when the yeast has accumulated around a tumor. Lastly, in order for the yeast to be able to report the presence of cancer cells, we make it express gas vesicles that can be detected with the help of ultrasound.
Organism: Saccharomyces boulardii
For our product to work we need to implement it in an organism that can survive in the gut of the patient, but that does not have pathogenic properties. Since the yeast Saccharomyces cerevisiae is commonly used in our lab the first organism of choice was yeast. However, S. cerevisiae is not adapted to the pH variations and higher temperatures in the human gut environment (Palma et al., 2015) and is therefore not the best candidate organism for our product. On the other hand the probiotic yeast Saccharomyces boulardii, a subspecies of S.cerevisiae, is better adapted to the gut pH variations and has an optimal growth temperature of 37 ° (Czerucka et al., 2007; Edwards-Ingram et al., 2007; Palma et al. 2015; Liu et al. 2016). S. boulardii already has a GRAS (Generally Regarded As Safe) status and the genetic makeup is very similar to that of S.cerevisiae, apart from differences in gene copy numbers (Edwards-Ingram et al., 2007; Liu et al., 2016). Since we are testing our system in S. cerevisiae in the lab, S.boulardii is a fitting target organism for our system. Right now several alternative probiotic S. cerevisiae strains are also under development, but none of these have a GRAS status at the moment (Palma et al.,2015). In the future, our system could also be implemented in these yeast strains.

Yeast to Cancer Cell Anchoring

In order to make it possible to locally treat the cancer, as well as to detect the tumor location, the yeast needs to bind specifically to the colon cancer cells. This is made possible through the use of Histone like protein A (HlpA) from the bacteria Streptococcus gallolyticus, which is commonly associated with colon cancer (Boleji et al. 2009; Ho et al., 2018). HlpA binds to the surface of the bacteria itself and also to heparan sulfate proteoglycans (HSPG), more specifically Syndecan 1 and possibly also Syndecan 2, on the surface of the colon cancer cell (Ho et al., 2018). In this way S. gallolyticus has been found to adhere to colon cancer cells and then infiltrate colon cancer tumors (Boleji et al. 2009).
Yeast surface anchor protein
In order to express HlpA at the S. boulardii cell surface an anchoring protein was needed since HlpA does not naturally adhere to the yeast cell surface. For this purpose, Alpha-Agglutin was used, since it is the most commonly used anchors' C-terminal anchor for expression of recombinant proteins at the yeast surface (Tanaka and Kondo, 2015). Alpha-Agglutin consists of two subunits, Aga1, and Aga2 (Tanaka and Kondo, 2015). Aga1 is expressed in the cell wall of the yeast, while Aga2 is secreted and then binds with the to Aga1 with a double disulfide bond (Tanaka and Kondo, 2015). In order to anchor HlpA at the yeast surface, the N-terminal of the protein was fused to the C-terminal of Aga2. A flexible linker was added in between the proteins, which should allow HlpA to be exposed at the yeast surface with the active site still functioning (Chen et at. 2013; Ho et al., 2018).
Cancer cell surface target proteins
As previously mentioned HlpA binds to HSPG’s present at the surface of the cancer cells. HSPG’s are in fact present at the surface of all epithelial cells in the gut, but usually, they are not exposed in a way that allows for the HlpA to bind effectively (Ho et al., 2018). However, in some cancer cell lines, there is a loss of apicobasal polarity, as well as changes in HSPG expression (Boleji et al. 2009; Ho et al., 2018). This results in Syndecan 1 and 2 being exposed at the cell surface, and thereby it allows for HlpA to bind (Ho et al., 2018). Since the HSPG’s in healthy cells are less exposed this results in adherence at a higher rate to the colorectal cancer cells compared to healthy cells.
Illustration of cell binding

Target molecule

Gas vesicles

Natural buoyancy
Gas vesicles are hollow proteins complexes that take cylindrical shapes of 45 to 200 nm diameter (Walsby, 1994). In nature, gas vesicles are produced by a variety of aquatic microorganisms to control their buoyancy in water. A well-conserved operon of more than 10 genes is responsible for the production of these vesicles. The two most important genes are the genes Gas Vesicle Protein A or its homolog B and Gas Vesicle Protein C (GvpA/B and GvpC). They form the rigid structure of the gas vesicles (fig with GvpA and GvpC, check where it is from), which is permeable to gasses but not liquids (Sivertse et al, 2010). Thus, gas from the cytoplasm will diffuse into the vesicles while other components will be held out. The other genes present in the operon are less known but are thought to be mainly involved in regulation, folding and gas vesicle assembly. These proteins do not appear in the final structure (Sivertse et al, 2010).
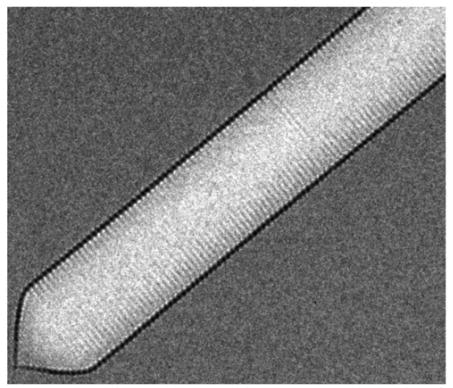
Acoustic reporter gene
An interesting property of these gas vesicles is that additionally to their original buoyancy function, they also refract ultrasound waves (Bourdeau et al., 2018). This property has been found to be useful in synthetic biology; Bourdeau et al. (2018) have shown that organisms can be engineered to produce gas vesicles as acoustic reporter genes. Because of their ultrasound scattering properties, gas vesicles can be observed non-invasively, even through non-transparent tissue, with ultrasound imaging instruments present in most modern hospitals. This could be done as represented in figure 3.2.

In their study, Bourdeau et al. (2018) have combined gas vesicle genes from Bacillus megaterium and Anabaena flos-aquae to produce the acoustic reporter gene in Escherichia coli as represented in figure 3.3.

Operon expression in yeast
In order to express this prokaryotic operon in the eukaryotic yeast, which is not able to transcribe gene operons, a multicistronic gene expression system is used. Namely, 2A viral peptides. This sequence, which we place in between each gene of the operon, codes for a peptide sequence that cleaves itself and separates the genes (Souza-Moreira et al. 2018). Without this system it would also be possible to express each gene separately, however, each gene should then be introduced under its own promoter and terminator while here, the gene set can be combined under one promoter and one terminator.

Our product
It has been mentioned before that our genetically modified probiotic yeast will target colon cancerous cells. Our idea is to put our yeast in a capsule that, passing through the gastrointestinal tract, will dissolve inside the colon releasing the microorganism. This delivery system has to be designed taking into consideration the gastrointestinal physiology (pH, microflora, enzymes, different fluid volumes and transit times) and also the increase of complexity when food is present. Together with these considerations, the inner part of the capsule should also provide a perfect environment for the yeast to survive and activate when it reaches the colon. The first idea would be to deliver the probiotic via rectal route because it is the shortest, however, it is difficult to reach the colon and patients could find it uncomfortable(Philip & Philip, 2010). Since we are trying to create a medicine as practical as possible we decided to opt for oral delivery route.PULSINCAP SYSTEM
For colon drug delivery, it can be useful to combine the effect of a PH sensitive and a time-released system. This is what Abraham et al. did in their Pulsincap system. This capsule is designed by three different parts and a coating:
- Water insoluble body: hard gelatin body treated with formaldehyde
- Hydrogel plug made of polymers such as: guar gum, hydroxypropylmethylcellulose 10K, carboxymethylcellulose sodium and sodium alginate(Abraham and Srinath, 2007)
- Water soluble cap: hydroxypropyl methylcellulose (HPMC)(‘Gastrointestinal-specific multiple drug release system’, 2005)
- Acid insoluble coating: 5% cellulose acetate phthalate

Combined with the capsule design, the probiotic yeast has to be prepared with a specific method and then placed inside of the capsule. We propose the same method that Hébrard G et.al used with S. boulardii: yeast is mixed with a 2:1 ratio of whey protein and alginate and then microparticles are formed through extrusion/cold gelation and then they are coated with WP or ALG by immersion(Hébrard et al., 2010).
References
Wang, Y., Wang, F., Wang, R., Zhao, P., & Xia, Q. (2015). 2A self-cleaving peptide-based multi-gene expression system in the silkworm Bombyx mori. Scientific Reports, 5(1), 16273. https://doi.org/10.1038/srep16273
Souza-Moreira, T. M., Navarrete, C., Chen, X., Zanelli, C. F., Valentini, S. R., Furlan, M., … Krivoruchko, A. (2018). Screening of 2A peptides for polycistronic gene expression in yeast. FEMS Yeast Research, 18(5). https://doi.org/10.1093/femsyr/foy036
Sivertsen, A. C., Bayro, M. J., Belenky, M., Griffin, R. G., & Herzfeld, J. (2010). Solid-State NMR Characterization of Gas Vesicle Structure. Biophysical Journal, 99(6), 1932–1939. https://doi.org/10.1016/J.BPJ.2010.06.041
Daviso, E., Belenky, M., Griffin, R. G., & Herzfeld, J. (2013). Gas Vesicles across Kingdoms: A Comparative Solid-State Nuclear Magnetic Resonance Study. Journal of Molecular Microbiology and Biotechnology, 23(4–5), 281–289. https://doi.org/10.1159/000351340
Walsby, A. E. (1994). Gas vesicles. Microbiological Reviews, 58(1), 94–144. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8177173
Philip, A., & Philip, B. (2010). Colon Targeted Drug Delivery Systems: A Review on Primary and Novel Approaches. Oman Medical Journal, 25(2), 70–78. https://doi.org/10.5001/omj.2010.24
Hébrard, G., Hoffart, V., Beyssac, E., Cardot, J.-M., Alric, M., & Subirade, M. (2010). Coated whey protein/alginate microparticles as oral controlled delivery systems for probiotic yeast. Journal of Microencapsulation, 27(4), 292–302. https://doi.org/10.3109/02652040903134529
Abraham, S., & Srinath, M. (2007). Development of modified pulsincap drug delivery system of metronidazole for drug targeting. Indian Journal of Pharmaceutical Sciences, 69(1), 24. https://doi.org/10.4103/0250-474X.32102
Tanaka, T., & Kondo, A. (2014). Cell-surface display of enzymes by the yeast Saccharomyces cerevisiae for synthetic biology. FEMS Yeast Research, n/a-n/a. https://doi.org/10.1111/1567-1364.12212
Ho, C. L., Tan, H. Q., Chua, K. J., Kang, A., Lim, K. H., Ling, K. L., … Chang, M. W. (2018). Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nature Biomedical Engineering, 2(1), 27–37. https://doi.org/10.1038/s41551-017-0181-y
Chen, X., Zaro, J. L., & Shen, W.-C. (2013). Fusion protein linkers: Property, design and functionality. Advanced Drug Delivery Reviews, 65(10), 1357–1369. https://doi.org/10.1016/j.addr.2012.09.039
Cheng, B., Montmasson, M., Terradot, L., & Rousselle, P. (2016). Syndecans as Cell Surface Receptors in Cancer Biology. A Focus on their Interaction with PDZ Domain Proteins. Frontiers in Pharmacology, 7. https://doi.org/10.3389/fphar.2016.00010
Palma, M. L., Zamith-Miranda, D., Martins, F. S., Bozza, F. A., Nimrichter, L., Montero-Lomeli, M., … Douradinha, B. (2015). Probiotic Saccharomyces cerevisiae strains as biotherapeutic tools: is there room for improvement? Applied Microbiology and Biotechnology, 99(16), 6563–6570. https://doi.org/10.1007/s00253-015-6776-x
Boleij, A., Schaeps, R. M. J., de Kleijn, S., Hermans, P. W., Glaser, P., Pancholi, V., … Tjalsma, H. (2009). Surface-Exposed Histone-Like Protein A Modulates Adherence of Streptococcus gallolyticus to Colon Adenocarcinoma Cells. Infection and Immunity, 77(12), 5519–5527. https://doi.org/10.1128/IAI.00384-09
Liu, J.-J., Kong, I. I., Zhang, G.-C., Jayakody, L. N., Kim, H., Xia, P.-F., … Jin, Y.-S. (2016). Metabolic Engineering of Probiotic Saccharomyces boulardii. Applied and Environmental Microbiology, 82(8), 2280–2287. https://doi.org/10.1128/AEM.00057-16
CZERUCKA, D., PICHE, T., & RAMPAL, P. (2007). Review article: yeast as probiotics -Saccharomyces boulardii. Alimentary Pharmacology & Therapeutics, 26(6), 767–778. https://doi.org/10.1111/j.1365-2036.2007.03442.x
Edwards-Ingram, L., Gitsham, P., Burton, N., Warhurst, G., Clarke, I., Hoyle, D., … Stateva, L. (2007). Genotypic and Physiological Characterization of Saccharomyces boulardii, the Probiotic Strain of Saccharomyces cerevisiae. Applied and Environmental Microbiology, 73(8), 2458–2467. https://doi.org/10.1128/AEM.02201-06
Elmer, McFarland, Surawicz, Danko, & Greenberg. (1999). Behaviour of Saccharomyces boulardii in recurrent Clostridium difficile disease patients. Alimentary Pharmacology and Therapeutics, 13(12), 1663–1668. https://doi.org/10.1046/j.1365-2036.1999.00666.x
Bourdeau, R. W., Lee-Gosselin, A., Lakshmanan, A., Farhadi, A., Kumar, S. R., Nety, S. P., & Shapiro, M. G. (2018). Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature. https://doi.org/10.1038/nature25021
Souza-Moreira, T. M., Navarrete, C., Chen, X., Zanelli, C. F., Valentini, S. R., Furlan, M., … De Montreal User, U. (2018). Screening of 2a Peptides for Polycistronic Gene Expression in Yeast, 46(May 2018). https://doi.org/10.1093/femsyr/foy036/4956763
