Careful planning puts you ahead in the long run; hurry and scurry puts you further behind
-- King Solomon
All these approaches require intense theoretical groundwork. Many hours have to be spent with screening literature, summarizing the current state of knowledge and finally, developing own ideas. Constructs have to be designed and experiments have to be planned.
All this preliminary work has to be completed before the lab work can be truly started. In addition to our initial theoretical work, we routinely iterated the steps: designing, building, testing and learning throughout the whole project.
On this page, we want to provide you the theoretical groundwork that we did to design our projects. We want to demonstrate that we followed Synbio design principles and conceived our project based on the current state of knowledge.
A major goal of our project was to create a collection of characterized parts for use in V. natriegens to provide other iGEM teams, as well as the whole scientific community a toolbox for the rational design of metabolic pathways, genetic circuits or any other DNA construct.
Our toolbox was initially conceived for being used in V. natriegens. However, while designing the Marburg Collection, we realized that a toolbox with maximum flexibility can easily be used in more than one bacterial species. Because alternative bacterial chassis, apart from E. coli, are gaining increasing importance (Kim et al. 2016), a toolbox that is compatible with more than one bacterial species enables scientists to work with organisms that have the exact properties needed for specific applications. Our vision is to establish the Marburg Collection as the first broad-host range golden-gate-based cloning toolbox.

De novo assembly of LVL1 plasmids from eight basic parts.
All basic parts like promoters or resistance parts are stored in LVL0 plasmids. The assembly of a plasmid comprising a single transcription unit is done by assembling at least eight parts resulting in one LVL1 plasmid. One to five LVL1 plasmids can then be used for a subsequent round of assembly to obtain a multigene LVL2 plasmid which could already harbor a full synthetic metabolic pathway consisting of up to five enzymes. Our toolbox even allows more rounds of assembly, each combining up to five constructs of the previous level. This enormous cloning capacity results theoretically in an infinite number of transcription units that can be assembled in a small amount of time.

Basic building blocks like promoters or terminators are stored in level 0 plasmids. Parts from each category of our collection can be chosen to built level 1 plasmids harboring a single transcription unit. Up to five transcription units can be assembled into a level 2 plasmid.
All LVL0 parts have to be stored in plasmids to allow for amplification and long term storage. To create new LVL0 parts, a PCR product or annealed oligos are cloned into a part entry vector. This vector harbours the resistance and ori that are required for selection and propagation. Furthermore, part entry vectors can be designed in a way that they contain a dropout. This dropout can be a transcription unit for a marker that generates a visible output. The first golden-gate-based toolbox MoClo (Weber et al. 2011) used a LacZ alpha transcription unit which can be used for blue white screening in many E. coli cloning strains. This concept was also adapted by iGEMs PhytoBrick system. During the cloning of LVL0 parts, this dropout is replaced by the desired part. When the cloning reaction is transformed into a suitable E. coli strain and the cells are plated on agar plates with supplemented IPTG and X-Gal. Colonies transformed with the religated entry plasmid appear blue while white colonies most probably contain the correctly assembled plasmid. The LVL0 part entry vector in iGEMs PhytoBrick system (BBa_P10500) has been designed as described and can be used for blue white screening.
We appreciate the approach of using part entry plasmids with dropouts but, for two reasons, we think that LacZ is not an optimal reporter. First, blue white screening requires the two expensive chemicals IPTG and X-Gal which have to be added to the agar plates. Second, blue white screening is restricted to E.coli strains with an incomplete lac operon that is complemented by the LacZ alpha fragment that is expressed from the plasmid (Langley et al. 1975). Consequently blue white screening is not compatible with a V. natriegens wild type strain (Link zu Improvement Page).
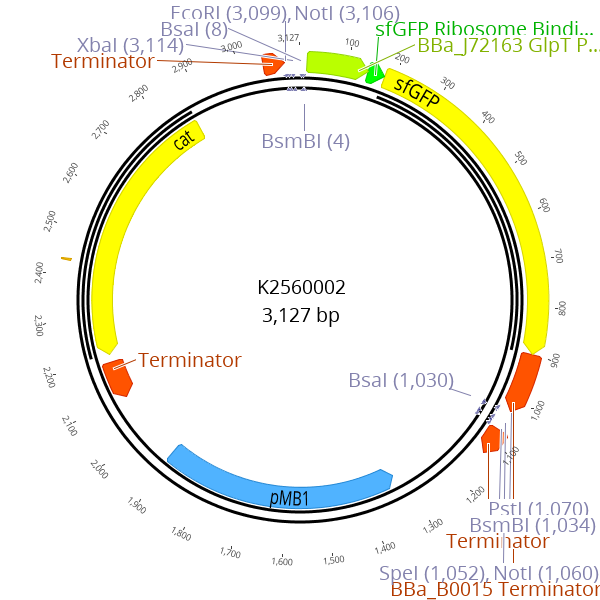
A sfGFP dropout cassette is flanked by BsmBI recognition sites. During the cloning of LVL0 parts with BsmBI, this dropout cassette is replaced by the desired part
We developed a solution for this problem by creating the novel resistance entry vectors BBa_K2560005 and BBa_K2560006. These plasmids contain a RFP and sfGFP dropout, respectively, and a chloramphenicol resistance cassette that is flanked by BsaI and BsmBI recognition sites. When a new LVL0 resistance part is cloned, the chloramphenicol resistance is replaced by the new antibiotic resistance marker resulting in a RFP or sfGFP expressing plasmid with the respective resistance marker. When using these LVL0 parts for LVL1 cloning, the re-ligated resistance parts yield colonies with a visually detectable phenotype. As a result, correct plasmids can be easily identified, even for inefficient LVL1 clonings with < 10 % efficiency.
Golden-gate based cloning relies upon the use of type IIs restriction endonucleases like BsaI, BsmBI or BpiI. In comparison to the commonly used type II restriction enzymes (e.g. EcoRI or PstI) they also recognize a specific DNA sequence but cleave outside of their recognition sequence (Pingoud and Jeltsch 2001). The golden-gate-cloning method is taking advantage of this property. A single enzyme can be used to create various single-stranded overhangs that match in a predefined order and finally lead to the correctly assembled plasmid (Engler et al. 2008).
When we started to design the Marburg Collection, we carefully investigated which fusion sites should be used. The fusion sites do not only set the order in which the single parts will be assembled but also affect the assembly efficiency and determine if a newly designed toolbox is compatible with already existing collections, so that parts can be shared easily. For us, the most important and decisive argument was that we wanted to be compatible with as many other toolboxes as possible. It is our strong belief that scientists all over the world should agree on one set of fusion sites to ensure complete interchangeability between different toolboxes. The toolboxes of MoClo (Weber et al. 2011), Loop Assembly (Pollak et al. 2018) and the PhytoBrick system already use a common set of fusion sites. We decided to adapt these fusion sites for all parts that build the transcription unit (Promoter, RBS, CDS, Terminator, Tags). Fusion sites for parts that are novel to our system (Connectors, Oris, Resistance cassettes) had to be newly designed by us because these parts did not exist in the other toolboxes.
We applied the following design principles to obtain optimal fusion sites. Firstly, the newly designed fusion sites must neither be identical to already existing fusion sites nor be palindromic to prevent assembly in a wrong order. Secondly, the fusion sites should not consist of bases that represent a portion of the recognition sequence of a restriction enzyme. If, for example, a fusion site with the sequence GGTC was used, and the sequence of the downstream part starts with TC, a BsaI recognition site would be reconstituted. So all fusion sites that would result in a partial recognition sequence of either BsaI, BsmBI or any of the enzymes that are used in the Biobrick cloning, are excluded. Lastly, the remaining candidates were sorted according to their GC content and the fusion sites with the highest GC content were chosen.
To make design of new parts as simple as possible, we created a collection of overhangs that can be copied from table xxxx and pasted to the sequence specific part of a primer to create new LVL0 parts. These primers contain the cut sites for integration into the part entry vector as well as the predefined fusion sites that are required for correctly assembling LVL1 plasmids. In some cases these overhangs contain additional bases that will be discussed in the following chapter.
Part Category
Fwd Overhang
Rev Overhang
1
5’ ConnectorAAGGTCTCGCTCGAACACGTCTCGNNNN
GGAGTGAGGGAGACCAA
2
PromoterAACGTCTCGCTCGGGAG
TACTTGAGGGAGACGAA
3
RBSAACGTCTCGCTCGTACTAGAG
TAATCAATGTGAGGGAGACGAA
4
CDSAACGTCTCGCTCGAATG
GCTTTGAGGGAGACGAA
5
TerminatorAACGTCTCGCTCGGCTTAA
CGCTTGAGGGAGACGAA
6
3’ ConnectorAAGGTCTCGCTCGCGCT
NNNNGGAGACGAGCTTGAGGGAGACCAA
7
OriAACGTCTCGCTCGAGCT
TGCTTGAGGGAGACGAA
8
ResistanceAACGTCTCGCTCGTGCTT
AACATGAGGGAGACGAA
4x
N-TagAACGTCTCGCTCGAATG
GGGATGTGAGGGAGACGAA
4y
N-tagged CDSAACGTCTCGCTCGGATG
GCTTTGAGGGAGACGAA
5a
C-TagAACGTCTCGCTCGGCTTTA
GGGTATGAGGGAGACGAA
5b
TerminatorAACGTCTCGCTCGGGTAA
CGCTTGAGGGAGACGAA
The fusion sites of most golden-gate-based cloning methods create a four base pair scar that is referred to as the fusion site. The fusion sites are the feature that makes toolboxes compatible with each other. Between some parts, additional bases are required for different reasons. These bases were chosen carefully to achieve best performance of the respective part. Please note that these additional bases are not a strict requirement to use or being compatible with our toolbox but we recommend them for the design of additional parts.
The first additional bases were incorporated between the Promoter and RBS part. The fusion site is TACT and AGAG was added additionally. The sequence between promoter and RBS that results by using our suggested overhangs form the same scar that is created if the the parts were assembled with 3A Assembly (Knight 2003). This means that the distance between promoter and RBS is not changed and therefore we do not expect negative effects in transcription or translation. Moreover, we hope that creating the same scar with a different method will make our experimental data more comparable to the data acquired with plasmids assembled with 3A Assembly in previous iGEM projects. The next bases were integrated between RBS and CDS parts. The fusion site, which was adapted from the PhytoBrick system, AATG and TAATC was added upstream of it. Previous work has shown that the sequence between a RBS and the start codon dramatically affects the expression of the desired protein (Lentini et al. 2013). A spacer length of six base pairs was shown to result in the strongest expression. When comparing different bases in a six bp spacer, the experimental data indicate significant differences. The 3A Assembly scar results in 50 % expression strength compared to the sequence TAATCT which was referred to as the reference (Lentini et al. 2013). We chose to use the spacer sequence which is expected to result in highest expression as we think that a system that is designed to enable strongest expression can be easily adapted for low expression by using weak RBS or promoters while going to the opposite direction might be more difficult. Unfortunately, we could not adapt the exact “reference sequence” because the first A in the fusion site AATG is already part of the spacer. Eventually, we used the first five bases of the strongest spacer (Lentini et al. 2013) upstream of the fusion site.
Close attention has to be paid to fusion sites that connect two sequences which are translated like the CDS part or N- and C- terminal Tags. A fusion site in our system consists of four bases which would result in disrupting the triplet code. To prevent mistranslated proteins, two additional bases have to be added to create a six bp scar that results in two translated amino acids. These can be seen as linkers between the joined CDS parts or tags. We decided to preferably use amino acids that are abundant in natural or synthetic flexible linkers like glycine and serine (Chen et al. 2012). Flexible linkers have been shown to improve the performance of epitope tags in Saccharomyces cerevisiae (Sabourin et al. 2007). Therefore we added the bases GG upstream of the fusion site between 4x (N-Tag) and 4y (CDS) which results in glycine and methionine (methionine is preset by the fusion site) and the bases TA downstream of the fusion site between 4 (CDS) and 5a (C-Tag) resulting in an alanine and leucine linker. To allow for the optional use of C-terminal tags, a CDS part must not possess a stop codon. Therefore an additional linker has to be introduced between 4 (CDS) and 5 (Terminator) as well as between 5a (C-Tag) and 5b (Terminator) resulting in an alanine-STOP and glycine-STOP, respectively.

Between some parts, additional base pairs were integrated to ensure correct spacing and to maintain the triplet code. We expanded our toolbox by providing N- and C- terminal tags by creating novel fusions and splitting the CDS and terminator part, respectively.
One key feature of the Marburg Collection are the connectors that provide our toolbox with the required flexibility. Our design is inspired by the “Dueber Toolbox”, a golden-gate-based cloning method designed for applications in Saccharomyces cerevisiae (Lee et al. 2015). To our knowledge, the “Dueber Toolbox” is the first cloning system that performs assembly through all levels using restriction sites located on independent basic parts, termed connectors, instead of destination plasmids for level 1 and level 2 plasmids (Weber et al. 2011). Using connectors instead of destination plasmids enables the user to freely choose antibiotic resistance parts and oris. To achieve the flexibility to choose one out of four resistances and one out of three oris, like it is possible in our toolbox, twelve plasmids would be required for building LVL1 plasmids. For building LVL2 plasmids, LVL1 plasmids with different fusion sites are needed. In our system, this is also achieved through the connectors in contrast to the destination plasmids which are used in other cloning methods. In already existing toolboxes, the combinatorics that come with choosing oris and resistances would be multiplied by the number of required positional vectors which provide the fusion sites for LVL2 assembly. A toolbox with five possible positions, four resistances and three oris would require 60 LVL1 destination plasmids. To enable inversion of transcription units, a built-in feature in our toolbox, the previous number would be multiplied by two, resulting in 120 required LVL1 destination plasmids.
Unsurprisingly, this theoretical toolbox would not be convenient to be built and used. The Marburg Collection presents a novel approach that achieves maximum flexibility with a minimum of required components. We provide two sets of connectors. The first set, the “short connectors” solely provide the fusion sites for subsequent assembly. The fusion sites are identical to the ones that are used in building LVL1 plasmids to avoid having to design a complete new set of fusion sites. This also enables using LVL0 ori and resistance parts in LVL2 plasmids. Inversion of individual transcription units was also achieved by designing fusion sites that match in reverse order. The 5’ and 3’ connectors can be chosen independently to build a LVL1 plasmid. This allows the user to combine any number of LVL1 plasmids between one and five into a LVL2 plasmid without end linkers that are required in other toolboxes (Andreou and Nakayama, 2018). In addition to the “short connectors”, we designed a set of “long connectors” that function as genetic insulators. They fulfill the same basic functions as the short connectors, providing fusion sites for subsequent cloning. Additionally, they consist of 300 bp “neutral DNA” flanked by two strong transcriptional terminators.

300 bp neutral DNA are flanked by two strong terminators. A BsmBI site for level 2 cloning is incorporated at the 5'end
We developed a Matlab script to find the “best” spacer sequences by increasing quantiles and picking spacers that fall into the lowest quantiles for all four categories. For example, the best sequence was found to be among the best 28 % of all tested sequences in terms of secondary structure, repeats, forbidden motifs and homology to the genome of V. natriegens. The selected sequences were flanked by synthetic transcriptional terminators that were developed in the lab of Christopher A. Voigt (Chen et al. 2013). Most terminators were described to be unidirectional (Chen et al. 2013) and therefore the orientation of the flanking terminators had to be considered. Our priority was to prevent transcription into the respective transcription unit from the upstream sequence. Therefore each spacer sequence was equipped with a strong terminator at the 3’ and 5’ end of the 5’ connector in forward and reverse orientation, respectively.
We expect to achieve the best reduction of crosstalk between neighbouring transcription units by combining our insulators in combination with connectors that facilitate inversion of an individual transcription unit, thus providing a large step towards the reliable and predictable design of synthetic circuits.

In addition to this standard set of parts, our toolbox provides N- and C-terminal fluorescence and epitope tags to facilitate fluorescence microscopy and protein purification and degradation tags that can be used to finetune protein levels. Lastly, we submitted all project related parts that can be used to perform genome engineering via CRISPR/Cas9 or natural competence and all enzymes that were used in our metabolic engineering project.
The Marburg Collection is designed to be the most flexible golden-gate-based toolbox for prokaryotes. The high degree of flexibility of our toolbox is achieved by the de novo construction of plasmids instead of using entry vectors like most other toolboxes. A novel core feature are the connectors that provide the fusion sites for subsequent cloning steps and can be used to set the order and orientation of each individual transcription unit. A set of newly designed insulators, consisting of “neutral DNA”, and two transcriptional terminators, are designed to minimize crosstalk between neighbouring transcription units.
The design of our biosensors was largely guided by the goal of having as little impact on the host metabolism, while simultaneously being simple and easy to implement. We chose to design all parts to be compatible with the Marburg toolbox, RFC10,RFC25,andRFC1000,
to make it as easy as possible for us and future users to implement it and adapt it to new requirements.

This part enables creation of new LVL0 parts by replacing the parts sequence with the desired insert. The sfGFP expression cassette is flanked by BsmBI recognition sites
Annotation of the location for the regulatory sequences upstream of the HdpR coding sequence was insufficient for us to precisely remove the activator binding site or the constitutive promoter driving HdpR expression. So, we included an about 200bp long region upstream of the HdpR, which should be enough to include all regulatory motifs. Also included is the HdpR coding sequence with its own ribosomal binding site (RBS). and promoter. This results in a part that includes the whole sensing function, but does not quite fit our scheme of clearly defined functional units. Since expression of any part cloned behind the regulatory region is directly controlled by it, and the part serves as both, a promoter and RBS. It is basically an inducible promoter fused to an RBS. Thus, we decided to give the part the overhangs of an RBS. on the downstream and a promoter on the upstream end.
In order to obtain the desired region from the P. putida KT2440 genome, we planned to amplify it via PCR and then isolate the fragment by gel purification of the corresponding band. Into the primer-design we factored in the overhangs we needed to make it Marburg toolbox compatible. So, we gave them non-binding overhangs which automatically added the necessary BsmBI cut sites.

This part enables creation of new LVL0 parts by replacing the parts sequence with the desired insert. The sfGFP expression cassette is flanked by BsmBI recognition sites
For the malonyl-CoA sensor, our proceeding was very similar. Just that we knew the exact location of all functional parts, except of the location of the promoter, form the 2015 paper by Liu et al and from annotations in the published genome (Liu et al. 2015). Therefore, we could implement it fully into the Marburg toolbox, by simply treating the FapR coding sequence as coding sequence. Not that straightforwardly was the design of the fapO regulator binding site. Since we did not know the promoter strength, and the mechanism of regulation was independent of the promoter sequence, we decided to implement a promoter from the Andersen library, J23100. By placing it right in front of the fapO site, we created a construct that, in theory, should give us a FapR controlled promoter. For the amplification of the individual parts from the B. subtilis 168 genome, again, we used primers sporting non-binding regions, adding the necessary overhangs for our toolbox.
For future projects, other sensors could also be employed. In a publication from 2016, Rogers et al. propose an alternative sensor for 3HP with a lower perception threshold (Rogerset al.2016). We choose not to implement it in our design because of its more complicated mechanism that requires the implementation of more genes. Intriguingly, it works by first converting 3HP to acrylate via a pathway of three heterologous enzymes. The acrylate is then sensed by a transcription factor, similarly to how our sensors described above work. They also report its successful implementation and application in increasing the 3HP titer up to 23-fold. It is conceivable that with a little more effort our system could yield comparable results. Off cause, s we have the additional benefit of having a much shorter iteration interval.
Liu, Di, Yi Xiao, Bradley S. Evans, and Fuzhong Zhang. 2015. Negative Feedback Regulation of Fatty Acid Production Based on a Malonyl-CoA Sensor-Actuator ACS Synthetic Biology 4(2): 132-40. http://www.ncbi.nlm.nih.gov/pubmed/24377365 (October 6, 2018).
Rogers, Jameson K., and George M. Church. 2016. Genetically Encoded Sensors Enable Real-Time Observation of Metabolite Production Proceedings of the National Academy of Sciences 113(9): 2388&-93. http://www.ncbi.nlm.nih.gov/pubmed/26858408 (October 13, 2018).

