Liposomes
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Esse beatae assumenda eaque ex recusandae pariatur sunt soluta modi facere laborum exercitationem odio iure magnam obcaecati quos voluptatibus placeat, ratione harum!
Provident, maxime ipsum veniam, rerum facere ad vero fugit ipsa natus recusandae sit voluptatum architecto laudantium vitae necessitatibus! Nesciunt illum porro sint odio sequi reprehenderit. Sint eligendi ex impedit recusandae!
Alias obcaecati impedit iure recusandae quas asperiores tempore sint, consectetur veniam provident iste nulla fugit velit aliquam expedita, assumenda repellat dolorem dolore! Sit quis dolorem ad pariatur repellat reiciendis officiis.
Asperiores molestiae eos quo inventore recusandae quae placeat delectus, natus sint. Ullam quas culpa nobis exercitationem omnis animi velit, deleniti fugit! Sapiente aperiam sit minima nostrum, rerum quae laudantium vero?
Qui blanditiis, excepturi veritatis eaque temporibus voluptate maxime facere laborum voluptatem rerum ex a ipsum voluptatibus tempore, saepe sunt omnis nostrum sint? Voluptatum facilis omnis ea accusantium explicabo magnam architecto!
Repellendus incidunt doloremque, a cum voluptates esse officia quia veniam architecto. Quibusdam deserunt nulla, dolore perspiciatis accusantium ad aliquam voluptatem iste iure, quae minus ipsa voluptates, sit voluptatum consequatur tempora?
Architecto impedit in repellendus, quo dolorum sequi voluptatum omnis maxime perspiciatis aspernatur obcaecati sint iusto tenetur praesentium labore. Natus eveniet quaerat recusandae dignissimos nam sed distinctio quos fugiat aliquid eligendi.
Molestias inventore nobis minus aspernatur recusandae asperiores excepturi ipsam voluptas quos, quae voluptatem voluptatibus vero veniam dolores fuga aliquid neque ut dolorem beatae cum temporibus tempora? Iure expedita debitis corrupti.
Quae quam earum impedit laborum, nobis aspernatur fugit enim consequuntur provident laboriosam obcaecati doloremque ipsam quis modi quos ratione ipsum beatae? Voluptas, debitis eum! Dolorem accusantium et rem. Veniam, dicta!
Sequi aut, eos id nemo maiores iste! Dicta cum eos, incidunt aperiam voluptate facilis vero vel deleniti inventore accusantium cupiditate saepe dolore atque quisquam voluptates aliquam amet a! Hic, consectetur.
Ribosome modifications
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Esse beatae assumenda eaque ex recusandae pariatur sunt soluta modi facere laborum exercitationem odio iure magnam obcaecati quos voluptatibus placeat, ratione harum!
Provident, maxime ipsum veniam, rerum facere ad vero fugit ipsa natus recusandae sit voluptatum architecto laudantium vitae necessitatibus! Nesciunt illum porro sint odio sequi reprehenderit. Sint eligendi ex impedit recusandae!
Alias obcaecati impedit iure recusandae quas asperiores tempore sint, consectetur veniam provident iste nulla fugit velit aliquam expedita, assumenda repellat dolorem dolore! Sit quis dolorem ad pariatur repellat reiciendis officiis.
Asperiores molestiae eos quo inventore recusandae quae placeat delectus, natus sint. Ullam quas culpa nobis exercitationem omnis animi velit, deleniti fugit! Sapiente aperiam sit minima nostrum, rerum quae laudantium vero?
Qui blanditiis, excepturi veritatis eaque temporibus voluptate maxime facere laborum voluptatem rerum ex a ipsum voluptatibus tempore, saepe sunt omnis nostrum sint? Voluptatum facilis omnis ea accusantium explicabo magnam architecto!
Repellendus incidunt doloremque, a cum voluptates esse officia quia veniam architecto. Quibusdam deserunt nulla, dolore perspiciatis accusantium ad aliquam voluptatem iste iure, quae minus ipsa voluptates, sit voluptatum consequatur tempora?
Architecto impedit in repellendus, quo dolorum sequi voluptatum omnis maxime perspiciatis aspernatur obcaecati sint iusto tenetur praesentium labore. Natus eveniet quaerat recusandae dignissimos nam sed distinctio quos fugiat aliquid eligendi.
Molestias inventore nobis minus aspernatur recusandae asperiores excepturi ipsam voluptas quos, quae voluptatem voluptatibus vero veniam dolores fuga aliquid neque ut dolorem beatae cum temporibus tempora? Iure expedita debitis corrupti.
Quae quam earum impedit laborum, nobis aspernatur fugit enim consequuntur provident laboriosam obcaecati doloremque ipsam quis modi quos ratione ipsum beatae? Voluptas, debitis eum! Dolorem accusantium et rem. Veniam, dicta!
Sequi aut, eos id nemo maiores iste! Dicta cum eos, incidunt aperiam voluptate facilis vero vel deleniti inventore accusantium cupiditate saepe dolore atque quisquam voluptates aliquam amet a! Hic, consectetur.
BAM complex
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Esse beatae assumenda eaque ex recusandae pariatur sunt soluta modi facere laborum exercitationem odio iure magnam obcaecati quos voluptatibus placeat, ratione harum!
Provident, maxime ipsum veniam, rerum facere ad vero fugit ipsa natus recusandae sit voluptatum architecto laudantium vitae necessitatibus! Nesciunt illum porro sint odio sequi reprehenderit. Sint eligendi ex impedit recusandae!
Alias obcaecati impedit iure recusandae quas asperiores tempore sint, consectetur veniam provident iste nulla fugit velit aliquam expedita, assumenda repellat dolorem dolore! Sit quis dolorem ad pariatur repellat reiciendis officiis.
Asperiores molestiae eos quo inventore recusandae quae placeat delectus, natus sint. Ullam quas culpa nobis exercitationem omnis animi velit, deleniti fugit! Sapiente aperiam sit minima nostrum, rerum quae laudantium vero?
Qui blanditiis, excepturi veritatis eaque temporibus voluptate maxime facere laborum voluptatem rerum ex a ipsum voluptatibus tempore, saepe sunt omnis nostrum sint? Voluptatum facilis omnis ea accusantium explicabo magnam architecto!
Repellendus incidunt doloremque, a cum voluptates esse officia quia veniam architecto. Quibusdam deserunt nulla, dolore perspiciatis accusantium ad aliquam voluptatem iste iure, quae minus ipsa voluptates, sit voluptatum consequatur tempora?
Architecto impedit in repellendus, quo dolorum sequi voluptatum omnis maxime perspiciatis aspernatur obcaecati sint iusto tenetur praesentium labore. Natus eveniet quaerat recusandae dignissimos nam sed distinctio quos fugiat aliquid eligendi.
Molestias inventore nobis minus aspernatur recusandae asperiores excepturi ipsam voluptas quos, quae voluptatem voluptatibus vero veniam dolores fuga aliquid neque ut dolorem beatae cum temporibus tempora? Iure expedita debitis corrupti.
Quae quam earum impedit laborum, nobis aspernatur fugit enim consequuntur provident laboriosam obcaecati doloremque ipsam quis modi quos ratione ipsum beatae? Voluptas, debitis eum! Dolorem accusantium et rem. Veniam, dicta!
Sequi aut, eos id nemo maiores iste! Dicta cum eos, incidunt aperiam voluptate facilis vero vel deleniti inventore accusantium cupiditate saepe dolore atque quisquam voluptates aliquam amet a! Hic, consectetur.
RNA Thermoswitches
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Esse beatae assumenda eaque ex recusandae pariatur sunt soluta modi facere laborum exercitationem odio iure magnam obcaecati quos voluptatibus placeat, ratione harum!
Provident, maxime ipsum veniam, rerum facere ad vero fugit ipsa natus recusandae sit voluptatum architecto laudantium vitae necessitatibus! Nesciunt illum porro sint odio sequi reprehenderit. Sint eligendi ex impedit recusandae!
Alias obcaecati impedit iure recusandae quas asperiores tempore sint, consectetur veniam provident iste nulla fugit velit aliquam expedita, assumenda repellat dolorem dolore! Sit quis dolorem ad pariatur repellat reiciendis officiis.
Asperiores molestiae eos quo inventore recusandae quae placeat delectus, natus sint. Ullam quas culpa nobis exercitationem omnis animi velit, deleniti fugit! Sapiente aperiam sit minima nostrum, rerum quae laudantium vero?
Qui blanditiis, excepturi veritatis eaque temporibus voluptate maxime facere laborum voluptatem rerum ex a ipsum voluptatibus tempore, saepe sunt omnis nostrum sint? Voluptatum facilis omnis ea accusantium explicabo magnam architecto!
Repellendus incidunt doloremque, a cum voluptates esse officia quia veniam architecto. Quibusdam deserunt nulla, dolore perspiciatis accusantium ad aliquam voluptatem iste iure, quae minus ipsa voluptates, sit voluptatum consequatur tempora?
Architecto impedit in repellendus, quo dolorum sequi voluptatum omnis maxime perspiciatis aspernatur obcaecati sint iusto tenetur praesentium labore. Natus eveniet quaerat recusandae dignissimos nam sed distinctio quos fugiat aliquid eligendi.
Molestias inventore nobis minus aspernatur recusandae asperiores excepturi ipsam voluptas quos, quae voluptatem voluptatibus vero veniam dolores fuga aliquid neque ut dolorem beatae cum temporibus tempora? Iure expedita debitis corrupti.
Quae quam earum impedit laborum, nobis aspernatur fugit enim consequuntur provident laboriosam obcaecati doloremque ipsam quis modi quos ratione ipsum beatae? Voluptas, debitis eum! Dolorem accusantium et rem. Veniam, dicta!
Sequi aut, eos id nemo maiores iste! Dicta cum eos, incidunt aperiam voluptate facilis vero vel deleniti inventore accusantium cupiditate saepe dolore atque quisquam voluptates aliquam amet a! Hic, consectetur.
Mistic fusion protein
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Esse beatae assumenda eaque ex recusandae pariatur sunt soluta modi facere laborum exercitationem odio iure magnam obcaecati quos voluptatibus placeat, ratione harum!
Provident, maxime ipsum veniam, rerum facere ad vero fugit ipsa natus recusandae sit voluptatum architecto laudantium vitae necessitatibus! Nesciunt illum porro sint odio sequi reprehenderit. Sint eligendi ex impedit recusandae!
Alias obcaecati impedit iure recusandae quas asperiores tempore sint, consectetur veniam provident iste nulla fugit velit aliquam expedita, assumenda repellat dolorem dolore! Sit quis dolorem ad pariatur repellat reiciendis officiis.
Asperiores molestiae eos quo inventore recusandae quae placeat delectus, natus sint. Ullam quas culpa nobis exercitationem omnis animi velit, deleniti fugit! Sapiente aperiam sit minima nostrum, rerum quae laudantium vero?
Qui blanditiis, excepturi veritatis eaque temporibus voluptate maxime facere laborum voluptatem rerum ex a ipsum voluptatibus tempore, saepe sunt omnis nostrum sint? Voluptatum facilis omnis ea accusantium explicabo magnam architecto!
Repellendus incidunt doloremque, a cum voluptates esse officia quia veniam architecto. Quibusdam deserunt nulla, dolore perspiciatis accusantium ad aliquam voluptatem iste iure, quae minus ipsa voluptates, sit voluptatum consequatur tempora?
Architecto impedit in repellendus, quo dolorum sequi voluptatum omnis maxime perspiciatis aspernatur obcaecati sint iusto tenetur praesentium labore. Natus eveniet quaerat recusandae dignissimos nam sed distinctio quos fugiat aliquid eligendi.
Molestias inventore nobis minus aspernatur recusandae asperiores excepturi ipsam voluptas quos, quae voluptatem voluptatibus vero veniam dolores fuga aliquid neque ut dolorem beatae cum temporibus tempora? Iure expedita debitis corrupti.
Quae quam earum impedit laborum, nobis aspernatur fugit enim consequuntur provident laboriosam obcaecati doloremque ipsam quis modi quos ratione ipsum beatae? Voluptas, debitis eum! Dolorem accusantium et rem. Veniam, dicta!
Sequi aut, eos id nemo maiores iste! Dicta cum eos, incidunt aperiam voluptate facilis vero vel deleniti inventore accusantium cupiditate saepe dolore atque quisquam voluptates aliquam amet a! Hic, consectetur.
Surface display system
ScFv Antibody
">
Background
scFv consists of a minimal functional antigen-binding domain of an antibody (~30 kDa) (Fig. 1) , in which the heavy variable chain (VH) and light variable chain (VL) are connected by Ser and Gly rich flexible linker. [1] In most cases scFv is expressed in bacteria, where it is produced in cytoplasm, a reducing environment, in which disulfide bonds are not able to form and protein is quickly degraded or aggregated. Although poor solubility and affinity limit scFvs’ applications, their stability can be improved by merging with other proteins. [2] When expressed in cell free system, scFv should form disulfide bonds with the help of additional molecules. Merging to a membrane protein would provide additional stability and would display scFv on liposome membrane, where its activity could be detected. These improved qualities make ScFv recombinant proteins a perfect tool to evaluate, if SynDrop system acts in an anticipated manner. Of all possible scFvs we decided to use scFv-anti vaginolysin, which binds and neutralizes toxin vaginolysin (VLY). Its main advantage is rapid (< 1 h) and cheap detection of activity by inhibition of erythrocyte lysis (Fig. 2). Looking into future applications, scFvs are also attractive targets of molecular evolution, because one round of evolution would last less than one day thus generating a and wide range of different scFv mutants. Those displaying the highest affinity for antigens could be selected and used as drugs or drug carriers.
 Fig. 1
Fig. 1 Simplified structure of scFv Antibody
 Fig. 2
Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
Results
scFv constructs were created BBa_K2622006"Kristina" and checked by colony PCR and DNA sequencing (link to Simas construct protocol"Kristina"). scFv synthesis was performed in a cell free system. Validation of protein expression was done by running a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), see (Fig. 3)
 Fig. 3
Fig. 3 SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.
Red arrows in the photo indicate scFv anti-vaginolysin (~27 kDa). As successful synthesis was confirmed, the next step was to check if protein folded correctly and was able to bind its antigen - vaginolysin. We examined this by erythrocyte-lysis test, which was performed by comparing erythrocytes incubated with VLY (erythrocytes burst open) and erythrocytes incubated with VLY that was previously incubated with scFv anti-vaginolysin (less or no erythrocyte lysis). Results revealed that scFv binded to vaginolysin and inhibited cell lysis. Graph in (Fig. 4) demonstrates that scFv indeed attenuated the lysis of erythrocytes. These result prove scFv activity in IVTT system.
 Fig. 4
Fig. 4 Percentage of erythrocyte lysis at different +/-scFv dilutions.
We then went one step further and constructed MstX-scFv_antiVLY (BBa_K2622038"Kristina") fusion protein, aiming to increase the stability of scFv having in mind future applications and experiments of exposing it on liposome surface. Fusion protein was expressed in E.coli cells; yellow to red arrows in (Fig. 5A) indicate MstX-scFv expression after induction with IPTG.
Finally, we expressed the protein in a cell free system (Fig. 5B) along with scFv in order to compare how well scFv accomplishes its function alone or binded to other protein. In this case MstX-scFv_antiVLY fusion did not show superior activity than scFv_antiVLY alone (Fig. 6). These results also reveal that scFv_antiVLY is very sensitive and loses its activity with time. Ist and IInd attempts were separated by 1-2 hours. This amount of time is enough to measure decreasing activity. This must be taken into account when performing future experiments.
 Fig. 5
Fig. 5 A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.
 Fig. 6
Fig. 6 Fig 6. Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.
Conclusions
We successfully expressed scFv_antiVLY antibody and MstX-scFv_antiVLY construct in E.coli cells as well as in a cell free system. We demonstrated that our scFv anti-vaginolysin can bind to its target antigen vaginolysin and inhibit erythrocyte-lysis reaction. Based in scFv general properties in IVTT activity, we conclude that scFvs are an elegant addition to SynDrop liposome display system.
Discussion
Small size of scFv makes it a widely researched antibody. It’s ability to penetrate deeply into tissues and trait to elicit low to none organism’s immune response, makes scFv the one of the best candidates for medical, diagnostic, and research applications [3]. Efficient and fast method for scFv generation is in demand. SynDrop liposome display system offers an ability to produce scFv in IVTT system and display them on membranes to facilitate rapid antigen binding. scFv on the other hand can also help us prove that our system work either cost efficiently or with extreme precision. scFv surface display is compatible with using fluorescence assisted cell sorting (FACS) to detect well functioning liposomes. [4] This would reduce amount of time needed for mutant sorting compared with enzyme-linked immunosorbent assay. Second, scFv display is compatible with the experiments described in this section. We have performed erythrocyte-lysis tests to prove the functional activity of scFv anti-vaginolysin that was synthesized in the IVTT system. Not all experiments with VLY indicated positive antibody activity. We hypothesized that proteins could have aggregated very quickly after IVTT expression and the amount of active antibody left in the solution was not enough to inhibit VLY in quantities, what would have been detectable. This hypothesis was further supported by several experiments, which revealed decreasing scFv antibody’s functional activity with time. Moreover, not every experiment was done just after IVTT reaction completed and spend few hours in +4 ˚C. Another option to test scFv, single or displayed on liposomes to gain most reliable results, is an ELISA test. It requires specific antibodies and tags (His-6x or Strep-tag) on scFv or MstX-scFv.
Refferences
- Monnier, P., Vigouroux, R. & Tassew, N. In Vivo Applications of Single Chain Fv (Variable Domain) (scFv) Fragments. Antibodies 2, 193-208 (2013).
- Wang, R. et al. Engineering production of functional scFv antibody in E. coli by co-expressing the molecule chaperone Skp. Frontiers in Cellular and Infection Microbiology 3, (2013).
- Ahmad, Z. et al. scFv Antibody: Principles and Clinical Application. Clinical and Developmental Immunology 2012, 1-15 (2012).
- Vorauer-Uhl, K., Wagner, A., Borth, N. & Katinger, H. Determination of liposome size distribution by flow cytometry. Cytometry 39, 166 (2000).

Fig. 1 A simplified mechanism of action of RNA thermometers. At lower temperatures the secondary messenger RNA (mRNA) stem-loop masks the ribosome binding site (RBS). Higher temperature induces melting of the hairpin which reveals the RBS to allow ribosome binding and initiation of translation.
Background
RNA thermometers are RNA-based genetic control tools that react to temperature changes1. Low temperatures keep the mRNA at a conformation that masks the ribosome binding site within the 5’ end untranslated region (UTR). Masking of the Shine-Dalgarno (SD) sequence restricts ribosome binding and subsequent protein-translation. Higher temperatures melt the hairpins of RNA secondary structure allowing the ribosomes to access SD sequence to initiate translation 1. In terms of applicability of RNA thermometers in in vitro systems, they display certain advantages over ribo- or toehold switches: they do not require binding of a ligand, metabolite or trigger RNA to induce the conformational change2,3, therefore are especially compatible with our liposome IVTT system. Keeping that in mind we have explored literature 1,4 and found five different RNA thermoswitches that we decided to test and build into our system in order to delay the translation of fusion construct bearing beta-barrel membrane protein. Furthermore, understanding the importance of expanding the library of well characterized and widely-applicable biobricks, we have de novo designed ( check RNA Thermoswitches model) six completely unique heat-inducible RNA thermometers.
Results
With custom IDT primers with overhangs bearing thermoswitch sequences we performed a PCR from a GFP gene containing plasmid and inserted RNA thermometers GJx (link: BBa_K2622010, BBa_K2622011, BBa_K2622012, BBa_K2622013, BBa_K2622014) upstream the GFP gene. Another set of primers was used to produce RNA thermometers Swx (link: BBa_K2622016, BBa_K2622017, BBa_K2622018, BBa_K2622019, BBa_K2622020, BBa_K2622021). PCR was successful and all products were the same size as expected for Swx constructs ~76 bp (Fig. 2). DNA gel electrophoresis was not performed for GJx constructs, because whole plasmid was multiplied and only 40-60 bp were inserted.
 Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
pRSET plasmid and Swx PCR products were digested with restriction enzymes and ligated, while GJx PCR products were phosphorylated and ligated to produce plasmids from linear products. DH5α competent cells were transformed and plated on lysogeny broth (LB) media with ampicillin (Amp) and grown for 16 hours. Positive colonies were selected by colony PCR or restriction analysis (Fig. 3 and Fig. 4) and grown in 5 mL LB media. Plasmids were purified and BL21 competent cells were transformed. Three tubes of every construct plus plasmid with GFP without RNA thermometer were grown till OD600 reached 0.4. Control samples were taken and protein expression was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG). One tube of every construct was grown in 24 ˚C, 30 ˚C, and 37 ˚C. Samples were taken after 1 and 2 hours. SDS-PAGE was run (for elaborate protocol see Notebook/Protocols). Fig. 5, Fig 6 and Fig. 7 depicts GFP expression at different temperatures. Although our RNA thermometers were designed to melt at 37 ˚C, some displayed leakiness to different extent. GJ3 (link:BBa_K2622011) RNA thermometer was the leakeast and allowed for GFP translation at lower temperatures. On the other hand, when grown at 37 ˚C, it unlocked the translation of GFP to highest yields. GJ2 (link:BBa_K2622010) was less leaky, but inhibited protein translation more strictly when grown at 37 ˚C. GJ6 (link: BBa_K2622012), GJ9 (link:BBa_K2622013), and GJ10 (link: BBa_K2622014) suppressed GFP production at 24 ˚C and 30 ˚C at similar level. They also inhibited translation to some extent at higher temperatures, meaning their melting temperature was not reached. Altogether these results prove, that our synthetic thermoswitches are temperature-responsive and act in physiological temperature range needed for IVTT reaction and also for BamA folding and membrane insertion.
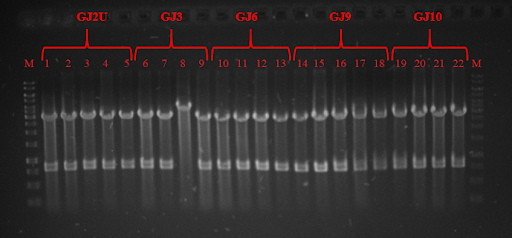 Fig. 3 Restriction analysis of GJx constructs
Fig. 3 Restriction analysis of GJx constructs
 Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
 Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
Discussion
As described in other sections of the Design and results page (check BAM Complex), beta-barrel bearing proteins are assembled into the membrane by the BAM protein complex machinery. The key protein BamA is itself a membrane protein, whose folding and insertion into membrane where it helps assemble target proteins, last up to two hours. In order to prevent the aggregation of our fusion proteins after encapsulating their gene-bearing plasmids and purified BamA mRNA into liposomes, we needed to develop a modulatory regulatory tool to lock the translation of our membrane proteins to allow enough time for the encapsulated BamA to fold and insert into the liposome membrane.
 Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
 Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.
Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.
Additionally, while creating SynDrop, we have considered various options on how to make our complex cell-free system more user-controllable and predictable. Cell-free systems are becoming an attractive platform for in vitro compartmentalization and protein research, and although usually compositionally sensitive, they also offer a platform for building synthetic genetic regulatory tools or logic gates. Both the need to control the translation time of target genes and desire to provide more modularity for our synthetic system, led us to exploring RNA thermometers as a viable option to perform these tasks. They have minimal molecular burden and are easy to modulate. These properties encouraged us to developed a library of synthetic RNA thermometers suitable to translationally regulate the expression of our fusion constructs in bacteria with a further possibility to transfer them to IVTT systems. All of the RNA thermometers including those we found in literature and our de novo modelled ones were optimized for best performance at 37oC, bearing in mind their future transition to IVTT system, whose optimum performance temperature is also 37oC. Consequently, our experiments showed that our synthetic RNA thermometers, despite their simplistic structures compared to naturally occurring ones, efficiently triggered the expression of target constructs at 37oC, and successfully locked it at lower temperatures having made them an ideal complement to our liposome IVTT system. All of our thermoswitches unlocked the expression to similarly high levels at 37oC, but differed in terms of leakiness and success at inhibiting translation at lower temperatures.
Conclusions
We proved that herein described synthetic RNA thermometers enable high-yield expression of our constructs in an inducible temperature range. What is more important, this spectrum of temperature is compatible with currently used in vitro transcription and translation systems. Synthetic Thermoswitches allow the user-controllable and responsive protein translation for custom experiments. Finally, we introduced six de novo designed RNA thermoswitches which will have been used by future iGEM teams having to work both with cell-free and in vivo synthetic biology systems.
References
- 1. Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. Oxford University Press; (2008); 36:e124–e124.
- 2. Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol. Rev. Wiley/Blackwell (10.1111); (2006); 30:3–16.
- 3. Storz G. An RNA thermometer. Genes Dev. Cold Spring Harbor Laboratory Press; (1999); 13:633–6.
- 4. Sen S, Apurva D, Satija R, Siegal D, Murray RM. Design of a Toolbox of RNA Thermometers. ACS Synth. Biol. (2017); 6:1461–70.


 Fig. 1 Simplified structure of scFv Antibody
Fig. 1 Simplified structure of scFv Antibody
 Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
 Fig. 3 SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.
Fig. 3 SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.
 Fig. 4 Percentage of erythrocyte lysis at different +/-scFv dilutions.
Fig. 4 Percentage of erythrocyte lysis at different +/-scFv dilutions.
 Fig. 5 A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.
Fig. 5 A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.
 Fig. 6 Fig 6. Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.
Fig. 6 Fig 6. Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.
 Fig. 1 A simplified mechanism of action of RNA thermometers. At lower temperatures the secondary messenger RNA (mRNA) stem-loop masks the ribosome binding site (RBS). Higher temperature induces melting of the hairpin which reveals the RBS to allow ribosome binding and initiation of translation.
Fig. 1 A simplified mechanism of action of RNA thermometers. At lower temperatures the secondary messenger RNA (mRNA) stem-loop masks the ribosome binding site (RBS). Higher temperature induces melting of the hairpin which reveals the RBS to allow ribosome binding and initiation of translation.
 Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
 Fig. 3 Restriction analysis of GJx constructs
Fig. 3 Restriction analysis of GJx constructs
 Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
 Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
 Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.
Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.