| Line 251: | Line 251: | ||
padding-left: 0.3em; | padding-left: 0.3em; | ||
"> | "> | ||
| − | <a href="https://static.igem.org/mediawiki/2018/ | + | <a href="https://static.igem.org/mediawiki/2018/9/90/T--Valencia_UPV--PCBRESULTSUPV2018.jpeg" |
data-lightbox="true"> | data-lightbox="true"> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/ | + | <img src="https://static.igem.org/mediawiki/2018/9/90/T--Valencia_UPV--PCBRESULTSUPV2018.jpeg" style=" |
margin-top: 0.5em;"> | margin-top: 0.5em;"> | ||
</a> | </a> | ||
| Line 259: | Line 259: | ||
<div style=" | <div style=" | ||
width: 100%; | width: 100%; | ||
| − | padding-left: | + | padding-left: 10em; |
padding-right: 1em; | padding-right: 1em; | ||
padding-bottom: 1em; | padding-bottom: 1em; | ||
Revision as of 23:16, 17 October 2018
Goals
At the start of the Printeria project, the different members of the Valencia_UPV team set themselves objectives in their respective areas in order to promote the automation of the cloning process and thus bring synthetic biology closer to people.
Wet Lab
Create a standardized Golden Gate basic Parts Collection to supply Printeria with the DNA toolkit to assemble composite parts.
Provide Printeria with the best way to transform competent bacteria with the highest transformation efficiency rate.
Avoid the plate colony screening step in order to eliminate the necessity of the researcher visual check, so easing Printeria automation.
Hardware
Integrate multiple lab equipment around an automated friendly form.
Design an easy way of handling a big variety of input liquids providing a straightforward integration with Printeria.
Design an easy to use way of handling a big variety of input liquids with easy integration of the machine.
Keep the footprint of the machine as small as possible.
Allow for maximum flexibility on the use of the machine.
Software
Develop a software that allows the user to control the device.
Allow the user to design a transcriptional unit with the software.
Allow the user to design multi-genetic constructs with the software.
Get non-professional users to print basic genetic circuits.
Fill out a “recipe” repository with at least 20 recipes.
Make a user friendly interface to interact with the software.
Control the device’s inventory through the software.
Simulate the bacterial growth and the protein expression.
Modeling
Design simple mathematical models based which allows us to simulate the different Printeria genetic circuits.
Optimize model parameters to match simulation results to experimental data obtained from Printeria.
Characterize the parts of our Part Collection from the optimization results and provide the user with all the information about the Printeria kit.
Develop a Simulation Tool that allows the user to visualize a prediction of the results of their experiment before running it in Printeria.
Results
Once the project was finished, the conclusions reached after our experiments were the following:
Wet Lab
We have built a complete Part Collection of standardized Golden Gate basic parts. It is a fully functional DNA collection with extensive characterization of its parts, so providing the user with useful data to work with.
We found that the most suitable way to transform bacteria for Printeria is to electroporate electrocompetent cells previously stored at -20ºC and subsequent maintained at 4ºC during the assembly step.
We proved that the assembly of composite parts using the destination vector in its linearized form significantly reduces the number of bacteria carrying the non-recombinant plasmid (false-positives colonies). However, in order to obtain a pure culture, it will always be necessary to go through the screening plate step.
Hardware
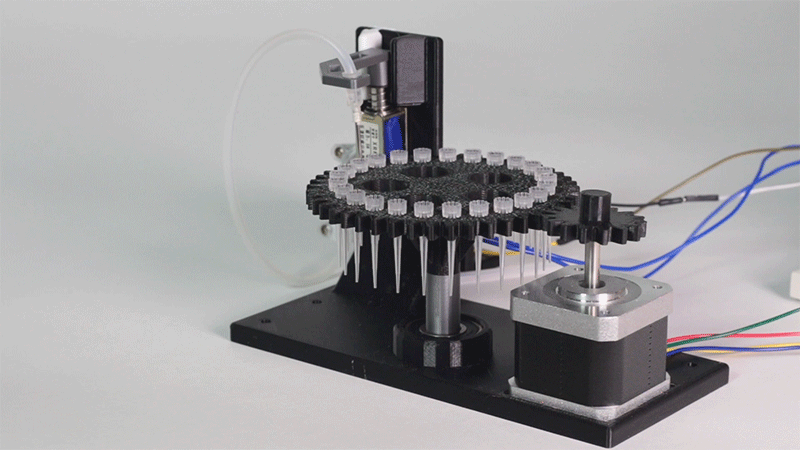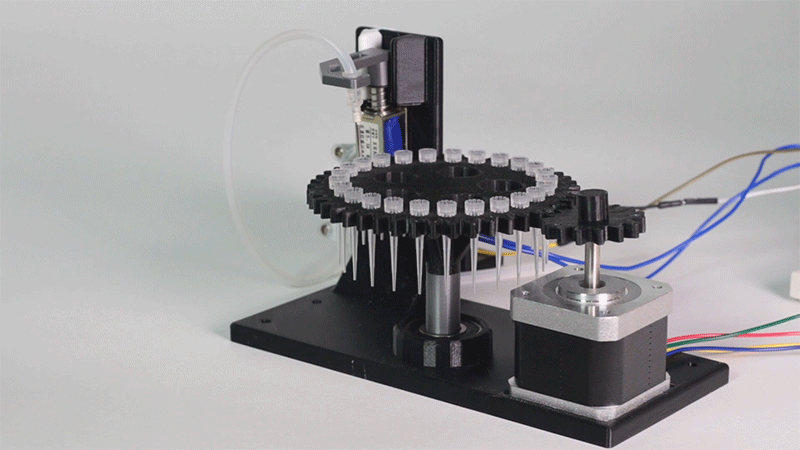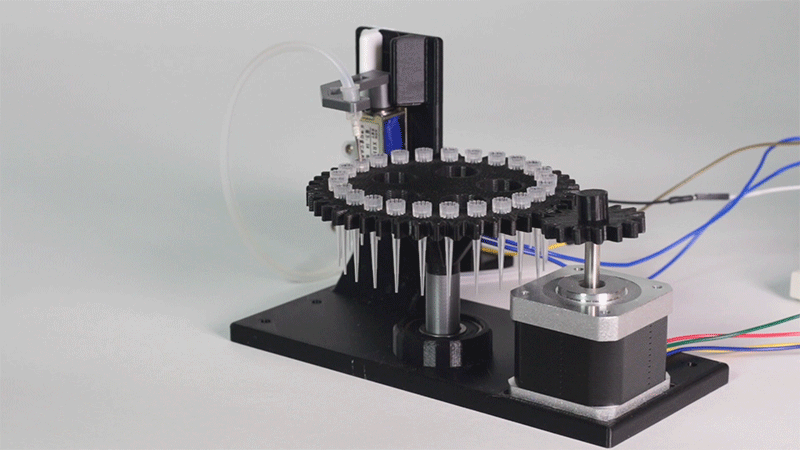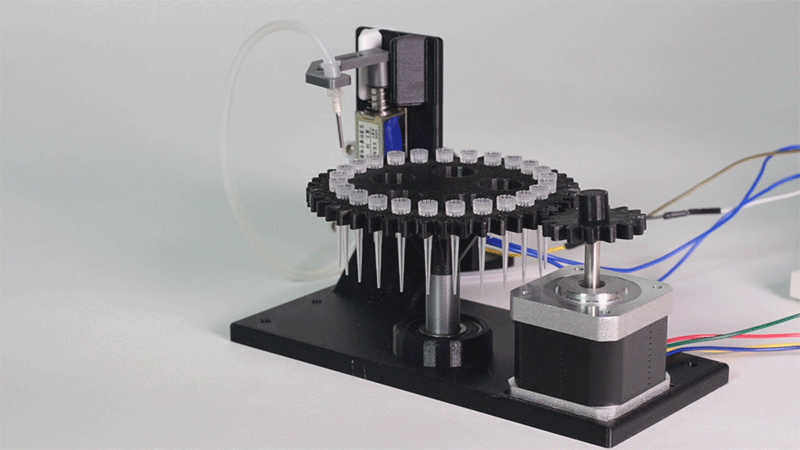
Laboratory equipment has been designed around digital microfluidic technology. Giving the machine a great degree of control over the processes that occur inside. This equipment includes a thermal cycler and an electroporator.
The entry system of Printeria allows for quick placement of all the necessary consumables. In addition, it can be further customized for any kind of liquid.
Thanks to the use of Digital Microfluidics Printeria can produce all the reactions in a small space.
All parts of the machine have been designed with individual use in mind.
Sensors have been included to monitor bacterial growth.
A functional, economical and easy-to-use solution has been achieved.
Software
We developed Printeria Controller, a web application capable to control all the functions of our device.


Image 4-5. Printeria controller software interface
Through the application it is possible to select all the DNA parts needed to design a transcriptional unit.
We didn’t implement the multi-genetic circuit constructions because the team considered this construction will be part of the future of printeria.
A non-experienced user can print a genetic circuit already created and stored in our recipe repository.


Image 6-7. Recipes and Parts interface of Printeria controller.
We created a database with 27 recipes assembled with our basic parts using the Golden Gate technology.
The clean and explicative interface of our software allows both experienced and non-experienced users to navigate through all the options.
In the Inventory tab of the software it is possible to know which cartridges have been drained and register when they are filled.
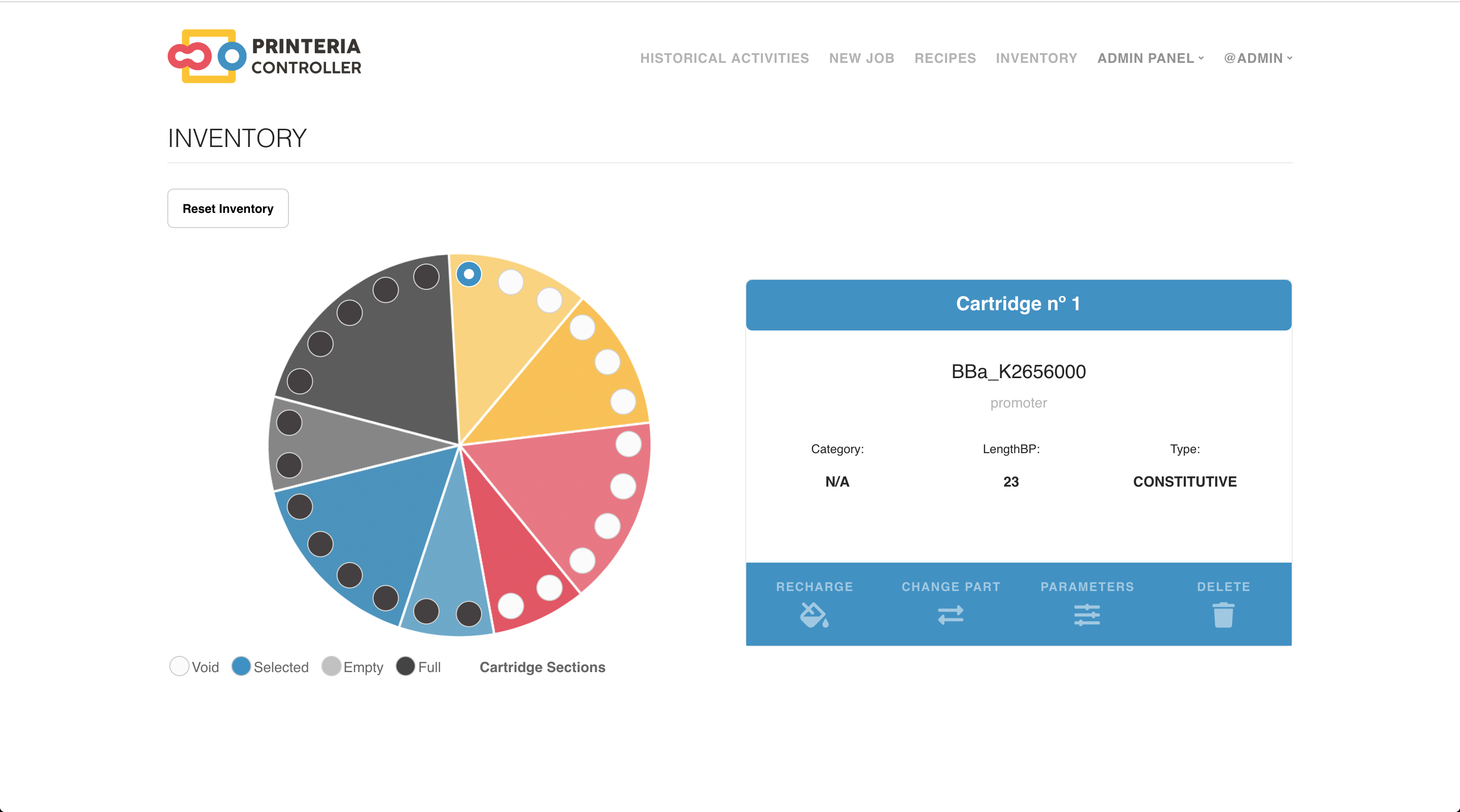
Image 8. Inventory interface of Printeria controller.
With the modeling team we developed a simulation tool that allow us to predict the results.
Modeling
Three types of mathematical models have been defined, with their scheme, reactions, parameters and equations: constitutive model, inducible model pBAD/araC and inducible model LuxR-LuxI.
The simulations of the theoretical models have been executed and the analyses have been carried out modifying the parameters of the model in order to determine its influence.
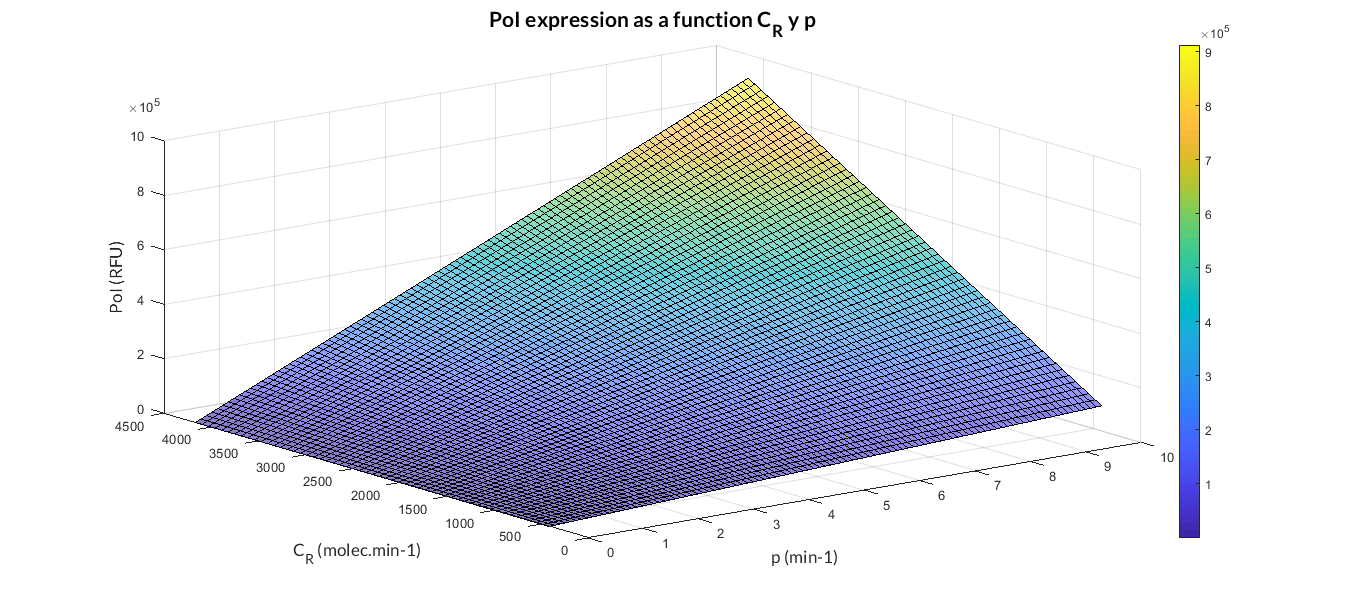
Constitutive expression model analysis. The graph represents de PoI stationary value as function of CR and p ratios. One example of how variable parameters influence the model.
We have established a protocol of experimentation in the laboratory, a process of correction and transformation of the data, and a protocol of multiobjective optimization of parameters.
Multiobjective experiments and optimization have been carried out satisfactorily with the constituent (changing promoters and RBS parts) and inducible LuxR-LuxI models and the optimal parameters of the parts have been estimated.
The spectra of the different reporter proteins and the equivalence factors between fluorescence per unit of absorbance (FOD) and the equivalent molecules of fluorescein have been obtained in order to obtain a more exact and non-arbitrary estimation of the concentration of the protein inside the cell.
Through experiments and the application of protocols, relevant information has been associated with the registered parts.
The Simulation Tool has been developed in collaboration with Printeria Software team. That tool includes the mathematical models developed, and with it the user can predict the experimental results of a transcriptional unit just clicking a button.
Considerations for replicating the project
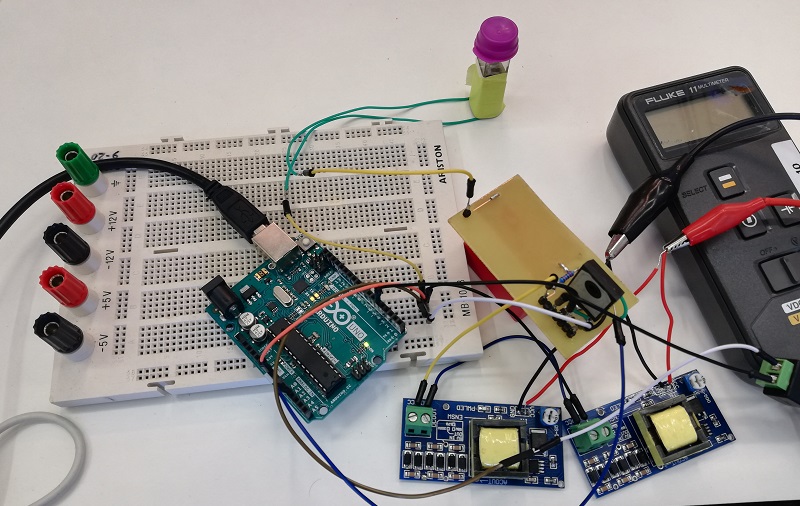
Special care: The electroporator has a capacitor at a really high voltage, act with care and avoid direct contact. You can isolate the PCB further with a layer of nail polish.
Ensure that if you use thermal paste on the peltier devices and the power resistor that is a non conductive paste.
For 3D Printed parts we used the standar settings for PLA, 0.15mm layer height, 40% infill, 215Cº.
The surface for the PCB is really hard to get right, give yourself some time to learn how to properly apply the coating.
While preparing the cables for the connection with the PCB make sure that all of them make good contact.
Remember that the experimental zone of the machine needs to be in complete darkness. We left the cover of that part transparent on the photos for display purposes.
When replicating Printeria biological function, take into account that the pre-defined assembly protocol is originally conceibed to be use with Golden Gate standandardized basic DNA pieces. Thus, you can easily use the pieces of our Part Collection , or you can adapt your desired pieces to the Golden Gate Technology.
Future plans
Until now, we have achieved the construction of transcriptional units, which is the basic level of assembly for any genetic circuit. However, our future aim is to finally achieve the contruction of multigenetical assemblies (Level 2), so increasing Printeria's application possibilities even more.
Moreover, we also want to reach the parallelization of several experiments. Currentlyy, we are limited with the design of the input and output zones to only three reactions. Thus, in the future the modification of this design, expanding the carrousel and PCB design, will allow us to do several more reactions at the same time.