Kristinazu (Talk | contribs) |
Kristinazu (Talk | contribs) |
||
| Line 140: | Line 140: | ||
</div> | </div> | ||
<p> | <p> | ||
| − | The genome modifications were then carried according to | + | The genome modifications were then carried according to <a href="https://2018.igem.org/Team:Vilnius-Lithuania/Protocols">Protocols</a> our protocol. Although cPCR gave us mixed results, we could not verify any colonies that afterwards grew on our selected marker antibiotics, and thus could not continue our experiments with them. It appears most likely that the genome modifications were not entirely successful, due to the somewhat unstable nature of the ligated linear DNA used for the donor sequence. |
</p> | </p> | ||
<p></p> | <p></p> | ||
| Line 214: | Line 214: | ||
<p></p> | <p></p> | ||
<p> | <p> | ||
| − | RNA thermometers are RNA-based genetic control tools that react to temperature changes<sup>1</sup>. Low temperatures keep the mRNA at a conformation that masks the ribosome binding site within the 5’ end untranslated region (UTR). Masking of the Shine-Dalgarno (SD) sequence restricts ribosome binding and subsequent protein-translation. Higher temperatures melt the hairpins of RNA secondary structure allowing the ribosomes to access SD sequence to initiate translation <sup>1</sup>. In terms of applicability of RNA thermometers in <var>in vitro</var> systems, they display certain advantages over ribo- or toehold switches: they do not require binding of a ligand, metabolite or trigger RNA to induce the conformational change<sup>2,3</sup>, therefore are especially compatible with our liposome IVTT system. Keeping that in mind we have explored literature <sup>1,4</sup> and found five different RNA thermoswitches that we decided to test and build into our system in order to delay the translation of fusion construct bearing beta-barrel membrane protein. Furthermore, understanding the importance of expanding the library of well characterized and widely-applicable biobricks, we have <var>de novo</var> designed (<a href= | + | RNA thermometers are RNA-based genetic control tools that react to temperature changes<sup>1</sup>. Low temperatures keep the mRNA at a conformation that masks the ribosome binding site within the 5’ end untranslated region (UTR). Masking of the Shine-Dalgarno (SD) sequence restricts ribosome binding and subsequent protein-translation. Higher temperatures melt the hairpins of RNA secondary structure allowing the ribosomes to access SD sequence to initiate translation <sup>1</sup>. In terms of applicability of RNA thermometers in <var>in vitro</var> systems, they display certain advantages over ribo- or toehold switches: they do not require binding of a ligand, metabolite or trigger RNA to induce the conformational change<sup>2,3</sup>, therefore are especially compatible with our liposome IVTT system. Keeping that in mind we have explored literature <sup>1,4</sup> and found five different RNA thermoswitches that we decided to test and build into our system in order to delay the translation of fusion construct bearing beta-barrel membrane protein. Furthermore, understanding the importance of expanding the library of well characterized and widely-applicable biobricks, we have <var>de novo</var> designed (<a href=https://2018.igem.org/Team:Vilnius-Lithuania/Model</a>) six completely unique heat-inducible RNA thermometers. |
</p> | </p> | ||
<p></p> | <p></p> | ||
| Line 253: | Line 253: | ||
<p></p> | <p></p> | ||
<p> | <p> | ||
| − | As described in other sections of the Design and results page (<a href= | + | As described in other sections of the Design and results page (<a href=https://2018.igem.org/Team:Vilnius-Lithuania/Design), beta-barrel bearing proteins are assembled into the membrane by the BAM protein complex machinery. The key protein BamA is itself a membrane protein, whose folding and insertion into membrane where it helps assemble target proteins, last up to two hours. In order to prevent the aggregation of our fusion proteins after encapsulating their gene-bearing plasmids and purified BamA mRNA into liposomes, we needed to develop a modulatory regulatory tool to lock the translation of our membrane proteins to allow enough time for the encapsulated BamA to fold and insert into the liposome membrane. |
</p> | </p> | ||
<p> | <p> | ||
Revision as of 23:17, 17 October 2018
Design and Results
Results
Cell-free, synthetic biology systems open new horizons in engineering biomolecular systems which feature complex, cell-like behaviors in the absence of living entities. Having no superior genetic control, user-controllable mechanisms to regulate gene expression are necessary to successfully operate these systems. We have created a small collection of synthetic RNA thermometers that enable temperature-dependent translation of membrane proteins, work well in cells and display great potential to be transferred to any in vitro protein synthesis system.


 Fig. 1 Principle of ribosome attachment to the liposome membrane. The ribosome exit tunnel is localized near the membrane, resulting in transmembrane domains of newly synthesized peptides interacting with the membrane, reducing aggregation
Fig. 1 Principle of ribosome attachment to the liposome membrane. The ribosome exit tunnel is localized near the membrane, resulting in transmembrane domains of newly synthesized peptides interacting with the membrane, reducing aggregation
 Fig. 2 Scheme of the genome modification process:
Fig. 2 Scheme of the genome modification process:
 Fig.3 Example of a constructed donor sequence. The sequence of the selected tag is present in primer used for the PCR of the homology arm that encompasses the target subunit. As a result, the tag sequence is fused to the ribosomal subunit gene.
Fig.3 Example of a constructed donor sequence. The sequence of the selected tag is present in primer used for the PCR of the homology arm that encompasses the target subunit. As a result, the tag sequence is fused to the ribosomal subunit gene.
 Fig. 4 PCR of homology arms, and antibiotic resistance genes
Fig. 4 PCR of homology arms, and antibiotic resistance genes
 Fig. 5 Constructed donor DNA sequences. The L29 donor DNA was not further revisited due to time constraints
Fig. 5 Constructed donor DNA sequences. The L29 donor DNA was not further revisited due to time constraints

 Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
Fig. 2 Electrophoresis gel of PCR products: 6 - Sw2, 7 - Sw3, 8 - Sw6, 9 - Sw7, 10 - Sw9, 11 - Sw11.
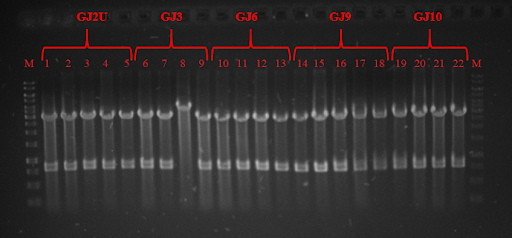 Fig. 3 Restriction analysis of GJx constructs
Fig. 3 Restriction analysis of GJx constructs
 Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
Fig. 4 Colony PCR of RNA thermometers in pSB1C3 plasmid.
 Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 5 expression at 24 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 6 GFP expression at 30 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
Fig. 7 GFP expression in 37 ˚C. On the right you can see GFP expression without RNA thermometer.
 Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
Fig. 8 Associational scheme of thermoswitches’ action in the SynDrop system. Not locking the concomitant translation of our target protein and BamA results in target protein aggregation due to insufficient membrane insertion and assembling potential of BamA.
 Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.
Fig. 9 Associational scheme of thermoswitches’ action in the SynDrop system. Locking up translation gives time for proper folding and insertion of BamA and prevents undesirable aggregation of target membrane proteins.




 Fig. 1 Simplified structure of scFv Antibody
Fig. 1 Simplified structure of scFv Antibody
 Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
 Fig. 3 SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.
Fig. 3 SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.
 Fig. 4 Percentage of erythrocyte lysis at different +/-scFv dilutions.
Fig. 4 Percentage of erythrocyte lysis at different +/-scFv dilutions.
 Fig. 5 A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.
Fig. 5 A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.
 Fig. 6 Fig 6. Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.
Fig. 6 Fig 6. Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.