
Optimizing a Cell-Free System
The first step of our project is the optimization of a cell-free expression system as a manufacturing platform for bacteriophages. For our purpose, it was necessary to produce a high-quality cell extrac, in a reproducible and easy manner. Considering a possible commercial use of our product we deemed it necessary to test the potential of upscaling of our preparation protocol.
We decided on the following optimization goals:
- increase protein content
- find reproducible methods of quality control
- test cell cultivation in a bioreactor to enable upscaling
- find optimal lysis conditions, that produce high-quality extract
- test upscaling of cell lysis
- produce cell-extract that allows phage assembly
Cultivation Can Be Upscaled In a Bioreactor
Cultivation is the first step in cell extract preparation. The original cell extract preparation protocol uses shaking flask cultivation for biomass production and states that cell harvest at OD 1,8-2,0 is strictly required to produce high-quality extract. To be able to transfer this to the bioreactor we first obtained growth data for both shaking flask and bioreactor cultivation.

The growth curve from shaking flask cultivation showed us that harvest at OD 1,8-2 correlates to the mid-to late logarithmic growth phase. In bioreactor fermentation this correlates to OD between 4 and 6. We decided to test which of these ODs is best to harvest cells for cell extract preparation.

This experiment gave 2 important results:
- the optimal OD to harvest culture from the bioreactor is around 5. This gives the highest protein content and also the best expression.
- preparing cell extract from bioreactor cultivated cells under comparable conditions gives equal quality extract than cell extract from shaking flask cultivated cells.
In our small lab-scale bioreactor with 2 L cultivation volume we were able to obtain 20 g cell pellet. 2 L cultivation in shaking flasks only yields 4,5 g pellet.
Sonication is The Superior Method For Cell Lysis
Of the three commonly used methods of cell lysis, each has their advantages and drawbacks. For cell extract preparation bead beating is most established, but the caveat there is, that it doesn’t leave room for upscaling, besides being tedious and time-consuming. Less often used is high-pressure cell disruption in a so called ‘french-press’, but these devices are very expensive and not widely prevalent which impedes our effort to provide a generally-applicable protocol for preparation of home-made cell extract. A sonication device on the other hand is available in most biochemical labs as it’s a widely used method for cell lysis in protein purification protocols. Nevertheless, we started by comparing all 3 lysis methods with settings commonly applied for cell lysis. For Sonication two commonly used sets of parameters were used.
Our Initial test showed that cell lysis by Sonication yields extract with up to 2,8 and 4,4 fold higher expression than cell lysis by bead beating and French press respectively. These findings, combined with the limited options of optimization for both bead beating and French press lysis as well as restricted potential for upscaling of bead beating lead to the decision to focus our optimization efforts on Sonication as Lysis method.
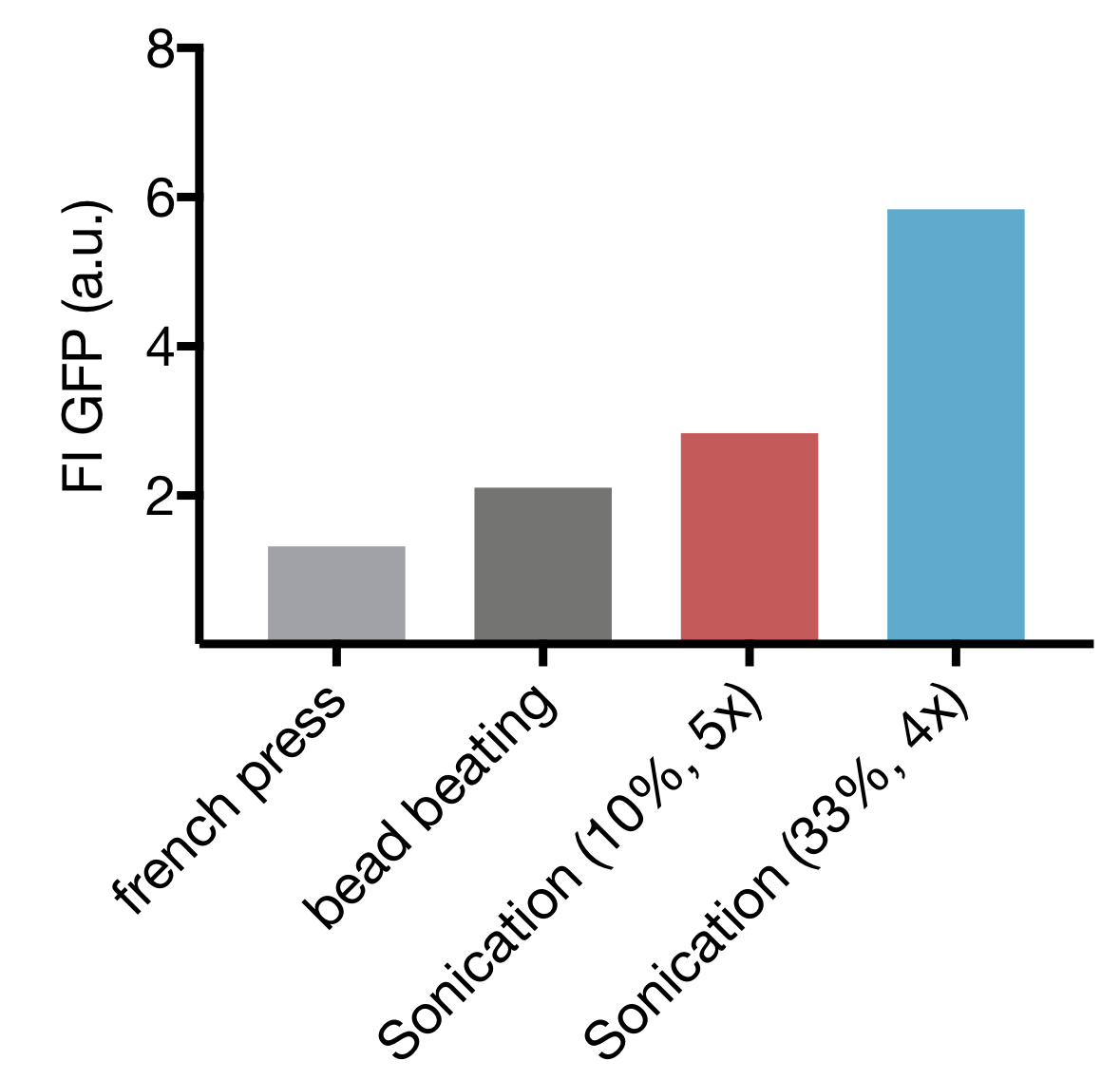
Comparison of different cell Lysis methods: Sonication at 33 % Amplitude, 4 cycles; Sonication at 10 % Amplitude, 5 cycles; bead beating and high-pressure disruption (french press) – Expression Quality in cell extract measured as GFP Fluorescence Intensity
Cell Lysis by Sonication offers 2 main parameters that can be varied: the Amplitude, that correlates to the energy emited by the device used and Cycle number, representing the total time of Sonication. AS mentioned in XY paper we decided to use 15 s pulses with 10 s pause between pulses to prevent excessive heating of the sample.
Our screening revealed that cycle numbers above 33 % seem to be inept for cell extract preparation, therefore our further efforts focused on Lysis with 10 % and 33 % amplitudes. Smaller cycle numbers offer the additional benefit of shorter overall processing time, cutting the cell expenditure of time for cell extract preparation short.

Lysozyme Improves Cell-Extract Quality
In the last step we made an effort towards upscaling of the sonication step. Preliminary experiments (results not shown) indicated 4 mL as the maximal sample volume that could efficiently be lysed with the device we had at our disposal. As indicated in the XY paper we decided to increase the number of sonication cycles in proportion to the increase of sample volume. The presented test revealed that this approach was successful; protein content in the cell extract and expression quality was not ustr maintained but increased through the use of higher sample volumes. This proves that upscaling of cell extract preparation with cell lysis by sonication is indeed feasible.


Additionally we tested the effect of addition of the cell-wall degrading enzyme Lysozyme to the cell lysis, following a tip from a lab member. To our knowledge usage of Lysozyme in cell extract preparation has not been a focus of previous studies. All the more surprised were we, to find that Lysozyme increases the protein yield of cell lysis for all tested settings. Moreover, the expression quality was likewise increased after Lysozyme addition, indicating that the remaining enzyme does not interfere with protein biosynthesis.
Post-Processing Of Cell-Free Systems Is Mandatory
The suggested steps in cell extract processing are:
- run-off reaction
- dialysis
- storage at -80°C
We found that the run-off reaction as suggested by Sun et.al is indeed beneficial and that dialysis is superior to diafiltration via centrifugal filters.
Quality Control



The TX-TL systems performed differentially regarding phage titers (E10>myTXTL>P15), protein content (E10>P15>myTXTL) and protein expression (P15>E10>Arbor), the proteome of the samples E10, P15 and myTXTL were analyzed by shotgun sequencing. The main question was, if the difference, especially in phage titers, is due to a different composition of the cell extracts or caused by a discrepancy in activity of the relevant proteins e.g. transcription and translation related proteins. Therefore, in collaboration with the Bavarian Biomolecular Mass Spectrometry Centre (BayBioMS) the samples were analyzed under supervision of Dr. Christina Ludwig. HEATMAP Heatmap of the label-free quantification intensities of all proteins identified. Diagram shows duplicate samples from the three cell extracts analyzed. Green indicates high expression, red indicates low expression.
The results of the Mass Spectrometry gave us insight into the composition of the different TX-TLs. We were able to identify 1771 proteins in all the extracts. The results indicate a high homogenicity between all three TX-TLs, as illustrated in the heatmap. Based on the LFQ values (Label Free Quantification) a volcano plot of two samples (Arbor/E10, Arbor/P15 and E10/P15) was generated.
VOLCANO In general, there is no protein more frequent in Arbor TX-TL in comparison to E10 and P15. In contrast, some translation-related proteins were slightly more abundant in E10 and P15, for instance certain tRNA ligases (PheS and PheT). The RecBCD subunits (the dsDNA degrading complex) are of similar abundance in all TX-TLs, showing the importance to consider its negative impact on the assembly. The results were further validated with DAVID, a tool for finding metabolic pathways based on proteomic data. The identified pathways indicate no correlation with transcription/translation related pathways (data not shown).
In conclusion, the performance differences are most likely caused by a loss of activity during TX-TL preparation rather than changes in composition.



































