(→Day 2) |
(→Day 2) |
||
| Line 332: | Line 332: | ||
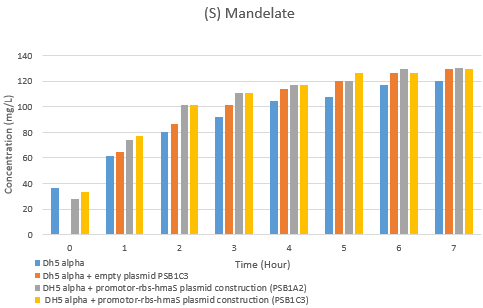
[[File:T--Aix-Marseille--Hplc s mandelate rate evolution.png|400px|right|]] | [[File:T--Aix-Marseille--Hplc s mandelate rate evolution.png|400px|right|]] | ||
<br> | <br> | ||
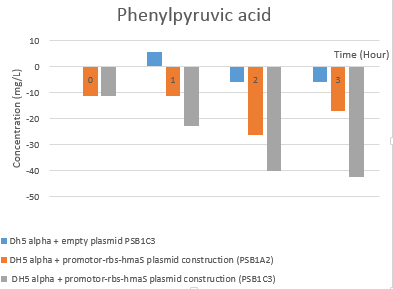
| − | [[File:T--Aix-Marseille--Hplc phenylpyruvic acid rate evolution new.png|400px| | + | [[File:T--Aix-Marseille--Hplc phenylpyruvic acid rate evolution new.png|400px|left|]] |
<br> | <br> | ||
Revision as of 21:54, 17 October 2018
Notebook
Contents
WEEK 1
Training
During the first week, we had several training sessions, under the supervision of Gauthier DANGLA-PELISSIER (Instructor), to get acquainted with the lab work, master everyday protocols (PCR/Colony-PCR, competent cells preparation, transformation, cloning, minipreps, PCR clean-ups...), and establish a classic workflow.
Day 1: Lab setup
We recovered equipment from our home university, cleaned-up the lab, and had a first meeting to establish the project's workflow.
Day 2: Preparing DH5α competent cells stock and transformation
We prepared a stock of DH5α competent cells. Afterward, we prepared Petri dishes and we transformed the cells to test their efficiency using a plasmid provided by our instructor.
Day 3: Colony-PCR and agarose gel electrophoresis
The next day we had colonies on our Petri dishes and wanted to check if they acquired the transformed plasmid with the right inserted gene. We picked out some supposedly positive colonies and ran a colony-PCR. We then migrated the PCR products on an agarose gel.
Day 4: DNA purification miniprep
We purified the plasmid with the right inserted gene from the confirmed positive colonies using a DNA purification kit. In a further step, we digested the purified plasmid with respective restriction enzymes to double-check the presence of the inserted gene.
Day 5: Western-blot
The last day, we checked the induced production rate of the relative protein to the inserted gene (Produced in a preliminary step by our instructor).
WEEK 2
For the whole week, we ran into a lot of problems making DH5α competent cells, so we spent a lot of time optimizing the protocol.
Day 1
Biobricks design: We designed Hmas, MdlB, MdlC and Hyd1 biobricks.
Megaprimers design: We designed megaprimers to add the Tag-his to our biobriks sequences.
Chitinase biobrick: We received the chitinase gene sequence from IDT and ran a PCR to amplify it.
Methionine-γ-lyase biobrick: We recovered the biobrick from the iGEM 2018 kit.
Day 2
pLacI promoter biobrick: We recovered the biobrick from the iGEM 2018 kit to assemble it with other biobricks.
Chitinase biobrick: We took the PCR product and verified the amplification on an agarose gel. We ran into hybridization problems so we had to determine the optimal hybridization temperature using a gradient-PCR.
Day 3
Chitinase biobrick: We cloned the PCR product in a pSB1C3 plasmid.
Methionine-γ-lyase biobrick: We transformed the biobrick (From the iGEM 2018 kit) into BL21 competent strains because we ran into problems making competent DH5α strains .
Day 4
Chitinase biobrick: We ran another cloning process on the previous PCR product and transformed it into DH5α competent cells.
Methionine-γ-lyase biobrick: We recovered the transformation plates and ran a colony-PCR on selected colonies. Afterward, we launched starters on the confirmed positive colonies.
Interlab: We had a training on the measurement devices and created the respective programs.
Biobrick design: We designed a new biobrick (OmpT-AIDAC) a cleavage sequence.
RFP biobrick: We recovered the biobrick (From the iGEM 2018 kit) that will be used for white-red screenings.
Day 5
pLacI promoter biobrick: We transformed the plasmid into DH5α competent cells.
Chitinase biobrick: We transformed DH5α strains with the plasmid containing the chitinase sequence.
Interlab: We recovered the 8 biobricks (From the iGEM 2018 kit) and transformed them into DH5α competent cells.
Methionine-γ-lyase biobrick: We purified the DNA using a DNA purification miniprep kit and stocked it.
MdlB biobrick: We received the biobrick and ran a PCR on it.
RFP biobrick: We transformed the biobrick into DH5α competent cells.
WEEK 3
Day 1
pLacI promoter biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
RFP biobrick: We recovered the transformation plates and launched overnight starters to purify the plasmid later on (No colony-PCR because the colonies are red: RFP expression).
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
We amplified the chitinase IDT gene sequence to recover the stock.
Methionine-γ-lyase biobrick: We recovered the methionine-gamma-lyase biobrick (from the iGEM 2018 kit) and transformed it into [Team:Aix-Marseille/Protocols#DH5α_competent_cells_preparation|DH5α competent cells]].
InterLab: We recovered the 8 transformations and launched 8 starters in duplicates.
Day 2
pLacI promoter biobrick: We purified the plasmid with the inserted RFP gene using a DNA purification kits, and restarted because the final DNA concentration was too low (13 ng/µL).
RFP biobrick: We purified the inserted RFP gene using DNA purification kits.
Chitinase biobrick: We ran a gradient-PCR to find the optimal primers hybridization temperature. We then extracted the DNA using a gel extraction kit, digested the gene with respective enzymes, and ran an overnight ligation in a pSB1C3 plasmid at 16°C.
DH5α competent cells: We prepared a new DH5α competent cells stock and verified their efficiency using an RFP plasmid.
Methionine-γ-lyase biobrick: We ran a colony-PCR on the positive colonies and verified it on an agarose gel.
InterLab: We launched the 3 calibration measurements and ran the first cell measurements.
Day 3
pLacI promoter biobrick: We purified the DNA using a different DNA purification miniprep kit.
Chitinase biobrick: We ran a ligation in pSB1C3 at room temperature.
DH5α competent cells: We launched DH5α starters to renew the competent cells stock.
Methionine-γ-lyase biobrick: We purified the DNA using the Promega DNA purification miniprep kit. Afterward, we launched an overnight
megapriming using specific primers to add a Tag-his to our gene sequence so we can purify the resulting protein later on.
InterLab: We analyzed the collected data and launched new starters to run a second measurement test.
MdlB biobrick: We ran a ligation in pSB1C3 at room temperature.
Day 4
Methionine-γ-lyase biobrick: We digested the magapriming product by DpnI (To cut the methylated GATC sites), and transformed it into DH5α competent strains.
DH5α competent cells: we renewed our stock and verified their efficiency using the RFP plasmid.
InterLab: We ran a second measurement test.
MdlB biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
Extracurricular: We made our breaking bugs logo using E. coli strains that express the GFP.
Day 5
DH5α competent cells: We double-checked the competent cells efficiency by transforming the RFP plasmid.
MdlB biobrick: We recovered the overnight starters, and purified the DNA using minpreps kits, measured the DNA concentration on the NanoDrop and ran a digestion test.
WEEK 4
We had the same problems making DH5α competent cells (contaminations...). We had the help of our instructor who pointed out a lot of flaws with what we were doing in the process.
Day 1
Chitinase biobrick: We took stocked strains and launched starters to purify the DNA the next day.
MdlB biobrick: We tried cloning the gene sequence into pSB1C3 and transformed the prouct into DH5α competent cells. Furthermore, we ran a PCR on the IDT sequence to renew our stock.
Interlab: We ran measurments for the third time and launched the CFUs protocol.
Methionine-γ-lyase biobrick: We sent the magrapriming product to sequencing to check if we managed to add the Tag-his.
Day 2
Chitinase biobrick: We purified the DNA using a DNA purification miniprep kit and had 225,87 ng/uL n concentration. Afterward, we sent the product for sequencing, and simultaneously, launched a megapriming to add the Tag-his for purification later-on.
RFP biobrick: We amplified it to renew our stock for contamination purposes.
Interlab: We had a training on how to do dilution series.
Methionine-γ-lyase biobrick: The sequencing results were positve: we managed to add the Tag-his to our sequence.
MdlB biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
Hyd1 biobrick: We received the gene sequence from IDT, ran a PCR on it and stocked it.
Day 3
Chitinase biobrick: We digested the megapriming product by DpnI and transformed it into DH5α competent cells.
MdlB biobrick: We tried cloning the gene sequence, for a second time, into pSB1C3 and transformed the product into DH5α competent cells. Furthermore, we ran a PCR on the IDT sequence to renew our stock.
Interlab: We launched starters for a second CFUs test.
Hyd1 biobrick: We migrated the PCR product on an agarose gel, and did a second PCR after a first failure. After a second failure, we ran a gradient-PCR to determine the correct hybridization temperature.
Day 4
Chitinase biobrick: We recovered the transformation plates. No colonies were observable whereas the sequencing results showed positive results (The chitinase sequence was inserted into the pSB1C3 plasmid. We suspected the DH5α cells: turns out they were Bacillus Subtilis strains (Smell check). We had to run another megapriming.
MdlB biobrick: We used the same strains that we used for the chitinase.
Interlab: We ran the CFUs test.
Hyd1 biobrick: We performed a PCR clean-up and migrated the PCR product.
Day 5
Chitinase biobrick: We digested the megapriming product by DpnI and transformed it into new DH5α competent cells.
Hyd1 biobrick: We cloned the gene sequence in pSB1C3 and trasnformed it into DH5α competent cells.
WEEK 5
Day 1
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We launched starters from the confirmed positive colonies for the next day.
Interlab: We counted the boxes and calculated the CFU/mL. Afterward, we prepared new Petri dishes to run another CFU test.
Hyd1 biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We had 8 confirmed positive colonies and we launched two starters from two of them for the next day.
MdlC biobrick: We received the gene sequence from IDT, then ran a PCR and a gradient-PCR on it, but we failed.
Methionine-γ-lyase biobrick: We ran a PCR on the pLacI and methionine-gamma-lyase fragments.
Hmas biobrick: We received the gene sequence from IDT and ran a PCR on it, but failed.
Day 2
Chitinase biobrick: We purified the DNA using a DNA purification miniprep kit and sent the product for sequencing.
Interlab: We made the dilution series and incubated the plates overnight.
Hyd1 biobrick: We purified the DNA using a DNA purification miniprep kit and digested it with specific restriction enzymes. We didn't have conclusive results, so we launched new different starters.
pLacI promoter biobrick: We launched a megapriming to add the RBS to the promoter.
Methionine-γ-lyase biobrick: We assembled the sequence with the pLacI promoter, tried to clone it in pSB1C3, and transformed the product into DH5α competent cells.
Day 3
MdlB biobrick: We tried cloning the gene sequence into pSB1C3 and transformed the product into DH5α competent cells.
Interlab: We counted the boxes and calculated the CFU/mL. We then elaborated a workflow for the flow cytometry measurements.
Hyd1 biobrick: We purified the DNA from a confirmed positive colony using a DNA purification miniprep kit.
pLacI promoter biobrick: We digested the magapriming product by DpnI (To cut the methylated GATC sites), and transformed it into DH5α competent cells.
Methionine-γ-lyase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we launched starters on the confirmed positive colonies.
Day 4
MdlB biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We then launched starters from the confirmed positive colonies.
Hyd1 biobrick: We sent the miniprep product for sequencing.
pLacI promoter biobrick: We launched starters on the confirmed positive colonies for the next day.
Methionine-γ-lyase biobrick: We purified the DNA using a DNA purification miniprep kit and sent the product for sequencing.
Day 5
Chitinase biobrick: We recovered the sequencing results which were negative.
pLacI promoter biobrick: We purified the DNA from a confirmed positive colony using a DNA purification miniprep kit and sent the product for sequencing.
WEEK 6
Day 1
Methionine-γ-lyase biobrick: We recovered the transformation plates and launched starters of the supposedly positive colonies with the good gene sequence (Assembled with pLacI).
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We prepared starters from the confirmed positive colonies.
Interlab: We prepared 16 starters from our GFP expressing strains for our flow cytometry measurements.
Day 2
MdlB biobrick: We cloned the gene in pSB1C3 and trasnformed it into DH5α competent cells.
Methionine-γ-lyase biobrick: We recovered the sequencing results that were negative (Sequence without pLacI). We to try assembling the pLacI biobrick to are gene sequence a second time.
Chitinase biobrick: We purified the DNA using a DNA purification miniprep kit and sent for sequencing.
Hyd1 biobrick: We recovered the sequencing results that were positive.
Interlab: We recovered our starters and took the fluorescence measurements on the TECAN and BIO-RAD S3e Cell Sorter flow cytometer.
Beauveria bassiana growth: We prepared a specific medium medium (Sabouraud) for the fungus so can start the growth process. We had liquid sabouraud stock and sabouraud, agar, and chloramphenicol Petri dishes.
Day 3
MdlB biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We had non-conclusive results.
Hmas biobrick: We cloned the gene sequence in pSB1C3 and transformed the product in DH5α competent cells.
Chitinase biobrick: We recovered the sequencing results that were negative (No Tag-his). We had to make another megapriming to try to add the tag-his on our gene sequencing with minor changes to the protocol.
Interlab: We retrieved and analyzed our measurements data.
Beauveria bassiana growth: We took aliquots and measured the OD600 to check-out the growth rate. Furthermore, we counted the cells on a Malassez cell.
Hmas biobrick: We cloned the gene sequence in pSB1C3 and transformed into DH5α competent cells.
Day 4
Hmas biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepare starters from the confirmed positive colonies.
Chitinase biobrick: We digested the megapriming product with DpnI and transformed it into DH5α competent cells.
Hmas biobrick: We recovered the transformation plates and ran colony-PCR on the supposedly positive colonies. We had no positive colonies.
Beauveria bassiana growth: We took aliquots and measured the OD600 to check-out the growth rate. Furthermore, we counted the cells on a Malassez cell.
Day 5
Hmas biobrick: We purified the DNA using a DNA purification miniprep kit and ran a digestion test to double-check if we had the gene inserted.
Chitinase biobrick: We recovered the transformation plates where we had no colonies. Another megapriming attent was launched.
Beauveria bassiana growth: We took aliquots and measured the OD600 to check-out the growth rate. Furthermore, we counted the cells on a Malassez cell.
WEEK 7
Day 1
MdlB biobrick: We tried optimizing the PCR protocol using different mixes because we were unable to amplify the gene properly. Nonetheless, we failed.
Lipase biobrick: Because IDT couldn't provide us with the gene sequence, we tried amplifying it from wild-type Beauveria bassiana strains.
Methionie-γ-lyase biobrick: We recovered the sequencing results. They were negative, the pLacI assembly to the gene sequence failed.
Chitinase biobrick: We digested the megapriming product with DnpI and transformed it in DH5α competent cells.
pLacI promoter biobrick: We launched a megapriming to add the RBS to the sequence.
Beauveria bassiana growth: We made dilution series on previously launched starters (At different temperatures 25°C, 30°C, and 37°C) to check out the growth rate of the fungus and calculate the CFU/mL.
Day 2
MdlC biobrick: We ran a gradient-PCR using the Phusion-HF DNA polymerase. We had non-conclusive positive results.
Methionie-γ-lyase biobrick: We tried assembling the to genes while cloning them (We digested pLacI with SpeI and PstI, and digested The tagged methionine-gamma-lyase sequence with XbaI and PstI.
Chitinase biobrick: We recovered the transformation plates but had no colonies. Afterward, we tried transforming the megapriming product for the second time and added an RFP control.
pLacI promoter biobrick: We digested the megapriming product with Dpn1 and transformed it into DH5α competent cells.
Hmas biobrick: We purified the DNA from an overnight culture, ran a digestion test and sent for sequencing.
Beauveria bassiana growth: We counted the cells using a Malassez cell.
Day 3
MdlC biobrick: We ran a PCR on the gene sequence and had positive results. Afterward, we purified the DNA using a DNA purification miniprep kit and had 70 ng/uL in concentration.
MdlB biobrick: We ran another gradient-PCR using the Phusion-HF DNA polymerase, and we increased the cycles' number. We had negative results.
Methionie-γ-lyase biobrick: We ran the ligation process and transformed the product in DH5α competent cells.
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we launched starters on the confirmed positive colonies.
pLacI promoter biobrick: We recovered the transformation plates and ran colony-PCR on the supposedly positive colonies. Afterward, we launched starters on the confirmed positive colonies.
Hmas biobrick: We recovered the sequencing results that were negative.
Beauveria bassiana growth: After 2 days of growth, we found out that the dilution series product are yeast.
Day 4
MdlC biobrick: We digested the sequence with EcoRI and PstI to clone it in pSB1C3 the next day.
Methionie-γ-lyase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we launched starters on the confirmed positive colonies.
Chitinase biobrick: We purified the DNA using a DNA purification miniprep kit and sent for sequencing.
pLacI promoter biobrick: We purified the DNA using a DNA purification miniprep kit and sent for sequencing.
Beauveria bassiana growth: We spent time debbugging the problem and tried to find a solution to eliminate the yeats.
Day 5
MdlC biobrick: We cloned the gene sequence in pSB1C3 and transformed it into competent cells.
MdlB biobrick: We ran another PCR on the gene sequence, but increased the matrix volume in the PCR mix. We had conclusive and positive results.
Methionie-γ-lyase biobrick: We purified the plasmid using a DNA purification miniprep kit and sent it for sequencing. We then digested the sequence with EcoRI and PstI to clone it in pSB1C3.
Beauveria bassiana growth: We recovered the plates growing at 25°C, where we observed preliminary filamentous colonies.
WEEK 8
Day 1
MdlC biobrick: We recovered the transformation plates where we had no colonies. We ran another digestion test on the MdlC gene sequence for cloning.
HmaS biobrick: We digested the gene sequence with EcoRI and PstI for cloning in pSB1C3. We had the right band at the right size when migrated on an agarose gel.
Chitinase biobrick: We ran a gradient-Megapriming test using AS-25 chitinase's primers.
Methionine-γ-lyase biobrick: We recovered the sequencing positive results. We managed to add the pLacI promoter to the gene sequence. We then digested our DNA with EcoRI an PstI to clone it in the pSB1C3 plasmid.
Day 2
MdlC biobrick: We cloned the digested gene sequence in a pSB1C3 plasmid and transformed it into DH5α competent cells.
We then ran a PCR to renew the DNA stock.
MdlB biobrick: We ran gradient-PCR using a Phusion DNA polymerase to determine the optimal primers hybridization temperature.
HmaS biobrick: We cloned the digested gene sequence in pSB1C3 and transformed into DH5α competent cells. Afterward, we ran a PCR to renew our DNA stocks but failed.
Chitinase biobrick: We digested the megrapriming product by DpnI and transformed it into DH5α competent cells.
Methionine-γ-lyase biobrick: We cloned our digested gene sequence in pSB1C3 and transformed it into DH5α competent cells.
Day 3
MdlC biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We then prepared starters from the confirmed positive colonies for plasmid purification.
MdlB biobrick: We cleaned-up the gradient-PCR product and digested the target plasmid. We had negative results when migrated on an agarose gel.
HmaS biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters on the confirmed positive colonies for plasmid purification.
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We then prepared starters from the confirmed positive colonies for plasmid purification.
Methionine-γ-lyase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies for plasmid purification.
Constructs for Beauveria bassiana: We searched for plasmids to transform our fungus. We found pEG202 and pJG4-5 (with prokaryotic and eukaryotic replication origins). Afterward, we trasnformed them into DH5α competent cells.
We designed the bar phosphinothricin resistance gene, a biobrick for fungus' growth selection.
Day 4
MdlC biobrick: We purified the plasmid using a DNA purification miniprep kit an sent samples for sequencing.
HmaS biobrick: We purified the plasmid and sent samples for sequencing.
Methionine-γ-lyase biobrick: We purified the DNA, transformed it into DH5α competent cells, and sent samples for sequencing.
Constructs for Beauveria bassiana: We recovered the trandformation plates and prepared starters for plasmid purification.
We recovered three biobricks from the iGEM 2018 kit: GPD eukaryotic promoter, eukaryotic IRES RBS, eukaryotic terminator. Afterward, we transformed them into DH5α competent cells.
Day 5
Chitinase biobrick: We purified the plasmid using a DNA purification miniprep kit an sent samples for sequencing.
Methionine-γ-lyase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
Constructs for Beauveria bassiana: We purified the plasmids using a DNA purification miniprep kit. To double-check the plasmids we digested pEG202 with NgoMIV and XbaI and pJG4-5 with AgeI and SpeI.
We recovered the transformation plates and ran colony-PCR for double-checking.
WEEK 9
Day 1
MdlC biobrick: We faced a lot of problems with the amplification from the IDT stock. We spent the week debugging and optimizing our protocol.
HmaS biobrick: We recovered the sequencing results. We were able to clone the gene. Afterward, we cloned it in another plasmid to add the pLacI promoter. We transformed the product into DH5α competent cells.
Chitinase biobrick: We ran another an exponential megapriming PCR to add a tag-his to our gene sequence. Afterward, we digested the PCR product we DpnI and transformed it into DH5α competent cells.
Beauveria bassiana growth: We prepared competent blastospores for future use, and stocked aliquotes at -80°C.
Day 2
HmaS biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies.
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies.
Day 3
HmaS biobrick: We purified the DNA plasmid using a DNA purification miniprep kit and sent samples for sequencing.
Chitinase biobrick: We purified the DNA plasmid using a DNA purification miniprep kit. Firstly, we digested the plasmid with XbaI and PstI to clone the gene in a plasmid with pLacI and RBS. Secondly, we digested the plasmid with NgoI and PstI to clone the gene in another plasmid with pLacI, RBS, and a signal peptide.
Day 4
HmaS biobrick: We recovered the sequencing results. We were able to clone the gene. Afterward, we cloned it in a pSB1C3 plasmid.
Chitinase biobrick: We cleaned-up the digestion products using the Machinery-Nagel kit. Afterward, we cloned the digested genes in respective plasmids and trasnformed the products into DH5α competent cells.
Day 5
MdlB biobrick: We received the sequence from IDT with an added tag-his. Then, we ran a PCR on it to amplify the gene.
Chitinase biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies.
WEEK 10
Day 1
Methionine-γ-lyase biobrick:
MdlC biobrick and MdlB biobrick: For the whole week, we spent a lot of time optimizing the PCR protocol (Double checking primer annealing, modfying the PCR program...).
HmaS biobrick: We transformed the pSB1C3 plasmid in BL21 strains to start the protein production using the following protocol.
Chitinase biobrick: We cloned the gene sequence in pLacI pSB1C3 plasmid and transformed into DH5α competent cells.
Day 2
Methionine-γ-lyase biobrick:
MdlC biobrick: After multiple clonings and transformations, we were finally able to send purified plasmid samples for sequencing.
MdlB biobrick: We kept optimizing portocols.
HmaS biobrick: We prepared starters according to the protocol. After the induced production of the protein, we ran a several western blots to check the production rates.
Chitinase biobrick: We recovered the transformation plates an ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies.
Day 3
Methionine-γ-lyase biobrick:
Chitinase biobrick: We purified the plasmid and sent samples for sequencing.
Day 4
Methionine-γ-lyase biobrick:
Chitinase biobrick: Werecovered the sequecning results. We have a full sequence.
Day 5
Methonine-γ-lyase biobrick:
WEEK 11
Day 1
Chitinase biobrick: We started the protein production using the following protocol. (It took 3 days).
Methionine-γ-lyase biobrick: We purified the produced protein.
HmaS biobrick: We calibrated the HPLCsystem in preparations to measurements to test the protein functionality.
MdlB biobrick: We recovered isolated colonies and ran an optimized colony-PCR. Afterward, we prepared starters from the positive colonies.
MdlC biobrick: We recovered the sequencing results. We had negatives ones, so we started over trying to optimize the protocols.
Day 2
Methionine-γ-lyase biobrick: We stored the purified samples at -20°C. Afterward, we created a standard curve to the test the functionality of the protein.
HmaS biobrick: We prepared starters to start the HPLC measurements.
MdlB biobrick: We purified the DNA and sent samples for sequencing.
MdlC biobrick: We digested the gene sequence with respective enzymes, cloned it in a pSB1C3 plasmid, and transformed it into DH5α competent cells.
Day 3
MdlC biobrick: We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive-colonies.
Day 4
Chitinase biobrick: After testing out different production conditions in DH5α strains, we stuck with the most optimal one to get to purify large quantities of the wanted protein.
MdlC biobrick: We purified the DNA and sent samples for sequencing.
Day 5
Chitinase biobrick: After testing out different production conditions in BL21 strains, we stuck with the most optimal one to get to purify large quantities of the wanted protein.
WEEK 12
Day 1
Methionine-γ-lyase biobrick : We tested for the first test with DTNB (5,5′-Dithiobis2-nitrobenzoic acid) on purified protein and absorbance measurement on the TECAN were made.
MdlC biobrick : We digested the gene sequence with their respective enzymes, cloned it into a pSB1C3 plasmid and extract it from the gel.
Chitinase biobrick: We we have purified and identified a large quantities of the wanted protein. We did on these proteins a blue revelation overnight with half bleaching solution and half Coomassie blue.
Day 2
Methionine-γ-lyase biobrick : We breaked cells with emulsiflex and made activity test on purified protein in triplicate with absorbance measurement on the TECAN.
MdlC biobrick : We transformed MdlC in PsB1C3 plasmid.
MdlB biobrick: We cloned it in a pSB1C3 plasmid, and transformed it in DH5α competent cells.
Chitinase biobrick: We prepared activity test solutions (réaction buffer) and discolored gels of the day before.
Promotor-RBS-peptide signal sequence ompA biobrick : We cloned RBS promotor and signal sequence in PSB1C3 plasmid and transformed them into DH5α competent cells.
Bar biobrick : We cloned RBS promotor and signal sequence in PSB1C3 plasmid and transformed them into DH5α competent cells.
Day 3
Methionine-γ-lyase biobrick : We made again activity test in triplicate with absorbance measurement on the TECAN. We start the analyse results.
MdlC biobrick : We ran a colony-PCR with Slic1 and 4 on the supposedly positive colonies. Afterward, we prepared starters from the confirmed positive colonies.
MdlB biobrick: We ran a colony-PCR with Slic 1 and 4 but we had negatives ones, so we started over trying to optimize the protocols.
Chitinase biobrick : We prepared all activity test solutions.
Promotor-RBS-peptide signal sequence ompA biobrick : We ran a colony-PCR but we had negatives ones, so we started over trying to optimize the protocols.
Bar biobrick : We ran a colony-PCR but we had negatives ones, so we started over trying to optimize the protocols.
Day 4
Methionine-γ-lyase biobrick : On cell culture of Methionine-γ-lyase needed for the activity test (DTNB test and MBTH test), we induced at OD 0,7 with IPTG and incubateur during 5h at 30°C. Measurement of OD was made after incubation and we freezed pellets at 20°C. We made then activity test with MTBH (3-Methyl-2-benzothiazolinone hydrazone hydrochloride hydrate) on purified protein and absorbance measurement was made on the TECAN but results were not good as expected.
MdlC biobrick : We recovered the transformation plates an ran a colony-PCR on the supposedly positive colonies of last week. Afterward, we prepared starters from the confirmed positive colonies.
MdlB biobrick: We ran a colony-PCR of last week but we had negatives ones, so we started over to optimize the protocols.
Chitinase biobrick : We made an etalon range to realised activity test at 30°C during one hour. 4466,58 ug/mL of chitinase (and contaminants) was obtained.
Day 5
Methionine-γ-lyase biobrick : We made same activity test with MTBH and absorbance measurement was made on the TECAN but results were not good.
MdlC biobrick : We purified the plasmid and sent samples for sequencing.
Chitinase biobrick : We made activity tests thanks to Schales'procedure, a colorimetric method for detecting reduced sugars released by chitin hydrolisis by chitinase.
WEEK 13
During the last week in the laboratory, we gathered all the results and analyzed them, and prepared our constructs for shipment to the iGEM headquarters. Afterward, we packed away our equipment and cleaned-up our 3 months workspace.