Elinramstrom (Talk | contribs) |
Elinramstrom (Talk | contribs) |
||
| Line 148: | Line 148: | ||
<p>By comparing how much the following methods agree with each other we can investigate if by using one approach or both, can help to reduce the variability in measurements between different labs. </p> | <p>By comparing how much the following methods agree with each other we can investigate if by using one approach or both, can help to reduce the variability in measurements between different labs. </p> | ||
| − | <h2>Conversion | + | <h2>Conversion Between Absorbance of Cells to Absorbance of a Known Concentration of Beads</h2> |
<p>By measuring the scattered light from a known concentration of silica beads that are roughly the same size and shape as a normal <i>E.coli</i> cells we converted each lab’s absorbance measurement into a standard “equivalent concentration of beads” measurement</p> | <p>By measuring the scattered light from a known concentration of silica beads that are roughly the same size and shape as a normal <i>E.coli</i> cells we converted each lab’s absorbance measurement into a standard “equivalent concentration of beads” measurement</p> | ||
| − | <h2>Counting | + | <h2>Counting Colony-Forming Units (CFUs) from the Sample</h2> |
<p>By spreading a known concentration of cells in liquid media on a plate to see how many colonies grow, we can determine the cell concentration of the sample as a whole since each colony should grow from one single cell. We determined the number of CFUs in negative and positive control samples to compute a conversion factor from absorbance to CFU. </p> | <p>By spreading a known concentration of cells in liquid media on a plate to see how many colonies grow, we can determine the cell concentration of the sample as a whole since each colony should grow from one single cell. We determined the number of CFUs in negative and positive control samples to compute a conversion factor from absorbance to CFU. </p> | ||
| Line 159: | Line 159: | ||
<h1>Material and Methods</h1> | <h1>Material and Methods</h1> | ||
| − | <h2>Conversion | + | <h2>Conversion Between Absorbance of Cells to Absorbance of a Known Concentration of Beads</h2> |
<p>We made three sets of unit calibration measurements: an OD600 reference point, a particle standard curve, and a fluorescein standard curve.<br><br> | <p>We made three sets of unit calibration measurements: an OD600 reference point, a particle standard curve, and a fluorescein standard curve.<br><br> | ||
| Line 311: | Line 311: | ||
| − | <h2>Method for | + | <h2>Method for Counting Colony-Forming Units (CFUs) from the Sample</h2> |
<p>To determine the CFU count, we counted colonies from two positive control (<a href="http://parts.igem.org/Part:BBa_I20270"><strong>BBa_I20270</strong></a>) plates and two negative control (<a href="http://parts.igem.org/Part:BBa_R0040"><strong>BBa_R0040</strong></a>) plates. The OD values of the liquid cultures of the four samples were measured and diluted in triplicates to OD = 0.1 in 1 mL of LB + Cam media. Each diluted sample was further diluted in series where samples with dilution factor 8 x 10<sup>-3</sup>, 8 x 10<sup>-4</sup> and 8 x 10<sup>-5</sup> was plated on LB + Cam plates.<br><br> | <p>To determine the CFU count, we counted colonies from two positive control (<a href="http://parts.igem.org/Part:BBa_I20270"><strong>BBa_I20270</strong></a>) plates and two negative control (<a href="http://parts.igem.org/Part:BBa_R0040"><strong>BBa_R0040</strong></a>) plates. The OD values of the liquid cultures of the four samples were measured and diluted in triplicates to OD = 0.1 in 1 mL of LB + Cam media. Each diluted sample was further diluted in series where samples with dilution factor 8 x 10<sup>-3</sup>, 8 x 10<sup>-4</sup> and 8 x 10<sup>-5</sup> was plated on LB + Cam plates.<br><br> | ||
| Line 319: | Line 319: | ||
| − | <h1>Our | + | <h1>Our Experience with the InterLab Study</h1> |
<p>The interlab is an excellent opportunity to practice cell transformations and streaking of plates. It was also very useful for us since we became more familiar with the iGEM kit plates and how to use the DNA provided in the starting kit. In addition we learned a lot from using the plate reader and the different calibrations, we gathered experience that became useful in other parts of our project.<br><br> | <p>The interlab is an excellent opportunity to practice cell transformations and streaking of plates. It was also very useful for us since we became more familiar with the iGEM kit plates and how to use the DNA provided in the starting kit. In addition we learned a lot from using the plate reader and the different calibrations, we gathered experience that became useful in other parts of our project.<br><br> | ||
Revision as of 23:08, 16 October 2018
Purpose
Synthetic biology is an emerging and constantly developing field which holds great promise to solve major problems humanity is facing today in terms of biology related medicine. In order to make use of this potential it is very important to reduce measurement variability and ensure there are standards in the field. To contribute towards this, our team has taken part in the iGEM interlab study, which has the noble goal of identifying and correcting the sources of systematic variability in synthetic biology measurement.
iGEM has in previous interlab studies showed that it is possible to reduce the variability in measurements between labs by measuring GFP expression in absolute fluorescence units which are calibrated against a known concentration of a chromoprotein. However, there is still a large source of variability in such measurements depending on the number of cells in the sample. By dividing the total fluorescence by the number of cells we can determine the mean expression level of GFP per cell. Usually, computing the optical density (OD) of the sample gets you an approximation of the number of cells present. OD measurements are however subject to high variability between labs due to spectrophotometer calibrations and is therefore not a precise approximation method. By instead using the direct method of cell count in each sample, one can remove this source of variability in measurements.
InterLab Study 2018
By comparing how much the following methods agree with each other we can investigate if by using one approach or both, can help to reduce the variability in measurements between different labs.
Conversion Between Absorbance of Cells to Absorbance of a Known Concentration of Beads
By measuring the scattered light from a known concentration of silica beads that are roughly the same size and shape as a normal E.coli cells we converted each lab’s absorbance measurement into a standard “equivalent concentration of beads” measurement
Counting Colony-Forming Units (CFUs) from the Sample
By spreading a known concentration of cells in liquid media on a plate to see how many colonies grow, we can determine the cell concentration of the sample as a whole since each colony should grow from one single cell. We determined the number of CFUs in negative and positive control samples to compute a conversion factor from absorbance to CFU.
Material and Methods
Conversion Between Absorbance of Cells to Absorbance of a Known Concentration of Beads
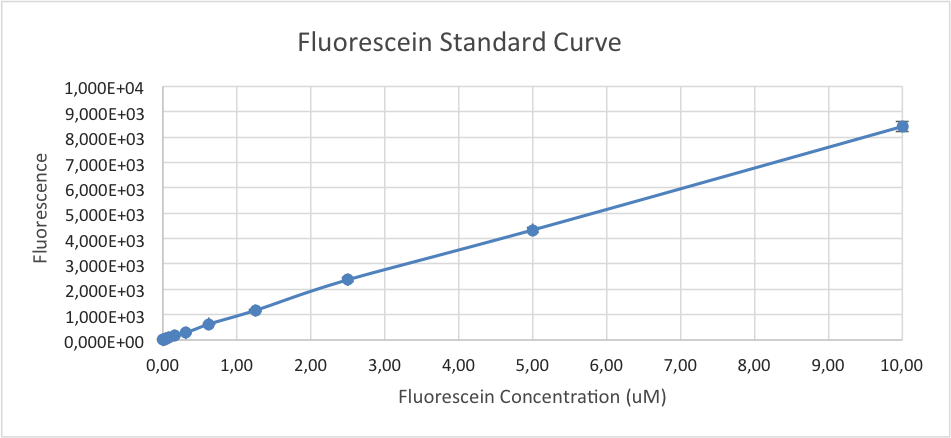
We made three sets of unit calibration measurements: an OD600 reference point, a particle standard curve, and a fluorescein standard curve.
Six different parts from the Distribution Kit along with a positive and negative control (all in a pSB1C3 backbone) were transformed into E. coli K-12 DH5-alpha cells. Fluorescence intensity and OD of the samples was measured with same the instrument and standard settings as the calibrations. Measurements were taken after 0 and 6 hours of liquid culture incubation in 37°C and 220 rpm.
Table 1: Parts and Devices used.
| Part | Part Number |
|---|---|
| Negative Control | BBa_R0040 |
| Positive Control | BBa_I20270 |
| Test Device 1 | BBa_J364000 |
| Test Device 2 | BBa_J364001 |
| Test Device 3 | BBa_J364002 |
| Test Device 4 | BBa_J364007 |
| Test Device 5 | BBa_J364008 |
| Test Device 6 | BBa_J364009 |
Figure 3: Transformation plates containing plasmids with different inserts, from the upper right to lower left: Test Device 4, Colony 2; Test Device 3, Colony 2; Test Device 1, Colony 2; Test Device 2, Colony 2.

Figure 4: Transformation plates containing plasmids with different inserts, from the right to left: Test Device 5, Colony 2; Test Device 6, Colony 2.

All plates were placed under a UV-light to confirm expression of GFP before picking colonies for inoculation. Fluorescent expression can be clearly distinguished from the expected cell culture, see figure 3 and figure 4.
Method for Counting Colony-Forming Units (CFUs) from the Sample
To determine the CFU count, we counted colonies from two positive control (BBa_I20270) plates and two negative control (BBa_R0040) plates. The OD values of the liquid cultures of the four samples were measured and diluted in triplicates to OD = 0.1 in 1 mL of LB + Cam media. Each diluted sample was further diluted in series where samples with dilution factor 8 x 10-3, 8 x 10-4 and 8 x 10-5 was plated on LB + Cam plates.
After Incubation at 37°C for 17 hours the colonies on the plates were counted. Based on the assumption that 1 bacterial cell gives rise to 1 colony, CFU per 1 mL of an OD = 0.1 was calculated by the formula:
#colonies x Final Dilution Factor = CFU/mL
Our Experience with the InterLab Study
The interlab is an excellent opportunity to practice cell transformations and streaking of plates. It was also very useful for us since we became more familiar with the iGEM kit plates and how to use the DNA provided in the starting kit. In addition we learned a lot from using the plate reader and the different calibrations, we gathered experience that became useful in other parts of our project.
We were however a bit confused when reading through the protocols. The protocol for conversion between absorbance of cells to absorbance of a known concentration of beads and the protocol for counting colony-forming units (CFUs) from the sample had different typos which made us believe that it was actually an appendix. In addition, the antibiotic used for selection in the plates (chloramphenicol) was written with different abbreviations between the two protocols; Cam and Chlor. This can be very misleading for an inexperienced student and/or a non-native english speaker. We would therefore recommend using the same abbreviations and formulations in all protocols included in the Interlab Study. In general the cell measurement protocols were quite confusing and could use some attention/optimization. The workflow could have been more clear and better formatted for the usage of a 96 well plate.
This year's Interlab Study gave us insight in where variability might arise in laboratory work. We strongly recommend all teams to participate in future interlab studies, not only for educational purposes but to also gather experience and become familiar with different measurements and methods. Lastly, it also feels good to be a part of a great interlaboratory project with a meaningful purpose.