(→Calibration 3: Fluorescence standard curve) |
|||
| (105 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | {{Aix-Marseille}} | + | {{Aix-Marseille|title=Interlab}} |
| − | + | ||
| − | + | ||
== Brief history == | == Brief history == | ||
| Line 10: | Line 8: | ||
== The fifth interlab study == | == The fifth interlab study == | ||
| − | The fifth international InterLab study aims to identify and correct the sources of systematic variability in synthetic biology measurements | + | The fifth international InterLab study aims to identify and correct the sources of systematic variability in synthetic biology measurements so that eventually, measurements that are taken in different labs will be no more variable than measurements taken within the same lab. This year, we helped in answering the following question: '''Can we reduce lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs) instead of OD?''' |
== Materials and methods == | == Materials and methods == | ||
| + | |||
| + | === Cell measurments === | ||
| − | For this study, we transformed six different plasmids, along with a negative and positive control into E. coli K-12 DH5-alpha. The plasmids, from here on referred to as devices, contain a constitutive promoter with low, medium or high strengths. The test devices express GFP (as a reporter in the pSB1C3 backbone). A plasmid containing a constitutive promoter and GFP sequence ([ | + | For this study, we transformed six different plasmids, along with a negative and positive control into E. coli K-12 DH5-alpha. The plasmids, from here on referred to as devices, contain a constitutive promoter with low, medium or high strengths. The test devices express GFP (as a reporter in the pSB1C3 backbone). A plasmid containing a constitutive promoter and GFP sequence ([http://parts.igem.org/Part:BBa_I20270 BBa_I20270]) and a plasmid containing a TetR repressible promoter ([http://parts.igem.org/Part:BBa_R0040 BBa_R0040]) in the pSB1C3 backbone, were used as positive and negative controls. All plasmids were delivered to us in the iGEM 2018 distribution kit. |
We made overnight cultures of two colonies of each transformation plate. The following day, the OD600 values of the overnight cultures were measured, diluted to a target OD600 of 0.02 and grown over 6 hours. Afterward, the absorbance (600 nm) and fluorescence (ex. 485 nm – em. 520 nm, gain = 50) of the growing cultures were measured in a 96 wells black plate in a plate reader (TECAN M200 Infinite) at 0, 3 and 6 hours. | We made overnight cultures of two colonies of each transformation plate. The following day, the OD600 values of the overnight cultures were measured, diluted to a target OD600 of 0.02 and grown over 6 hours. Afterward, the absorbance (600 nm) and fluorescence (ex. 485 nm – em. 520 nm, gain = 50) of the growing cultures were measured in a 96 wells black plate in a plate reader (TECAN M200 Infinite) at 0, 3 and 6 hours. | ||
| + | |||
| + | === Colony forming units (CFUs) === | ||
| + | |||
| + | This procedure can be used to calibrate OD600 to colony forming unit (CFUs) counts, which are directly relatable to the cell concentration of the culture (i.e. viable cell counts per mL). This protocol assumes that 1 bacterial cell will give rise to 1 colony. For the CFU protocol, we counted colonies for our two positive control ([http://parts.igem.org/Part:BBa_I20270 BBa_I20270]) cultures and our two negative control ([http://parts.igem.org/Part:BBa_R0040 BBa_R0040]) cultures. | ||
| + | |||
| + | We made overnight cultures. The following day, the OD600 values were measured and diluted to a target OD600 of 0.1. Afterward, we made a dilution series (5 dilutions) with our starting samples. Dilutions 3, 4 and 5 were incubated overnight on LB + agar plates. | ||
| + | |||
| + | Based on the assumption that 1 bacterial cell gives rise to 1 colony, colony forming units (CFU) per 1mL of an OD600 = 0.1 culture can be calculated following two steps: counting the colonies on each plate with fewer than 300 colonies, then multiplying the colony count by the final dilution factor on each plate. | ||
== Results == | == Results == | ||
| + | |||
=== Calibration 1: reference point === | === Calibration 1: reference point === | ||
| − | We measured Abs600 of LUDOX and H2O to use them as a reference point to obtain a ratiometric conversion factor. By multiplying this factor with the raw Abs600 measurement data, we could transform our absorbance data into a standard OD600 measurement, accounting for instrument differences. | + | We measured Abs600 of LUDOX CL-X and H2O to use them as a reference point to obtain a ratiometric conversion factor. By multiplying this factor with the raw Abs600 measurement data, we could transform our absorbance data into a standard OD600 measurement, accounting for instrument differences. |
| − | + | {| class="wikitable" | |
| + | ! | ||
| + | ! style="width: 100pt;" | LUDOX CL-X | ||
| + | ! H2O | ||
| − | === Calibration 2: | + | |- |
| + | | style="width: 100pt;" | Replicate 1 | ||
| + | | 0.0537 | ||
| + | | style="width: 100pt;" | 0.036 | ||
| + | |||
| + | |- | ||
| + | | Replicate 2 | ||
| + | | 0.054099999 | ||
| + | | 0.0359 | ||
| + | |||
| + | |- | ||
| + | | Replicate 3 | ||
| + | | 0.056299999 | ||
| + | | 0.0361 | ||
| + | |||
| + | |- | ||
| + | | Replicate 4 | ||
| + | | 0.056499999 | ||
| + | | 0.0361 | ||
| + | |||
| + | |- | ||
| + | | Arithmetic Mean | ||
| + | | 0.055 | ||
| + | | 0.036 | ||
| + | |||
| + | |- | ||
| + | | Corrected Abs600 | ||
| + | | 0.019 | ||
| + | | | ||
| + | |||
| + | |- | ||
| + | | Reference OD600 | ||
| + | | 0.063 | ||
| + | | | ||
| + | |||
| + | |- | ||
| + | | OD600/Abs600 | ||
| + | | 3.294 | ||
| + | | | ||
| + | |} | ||
| + | |||
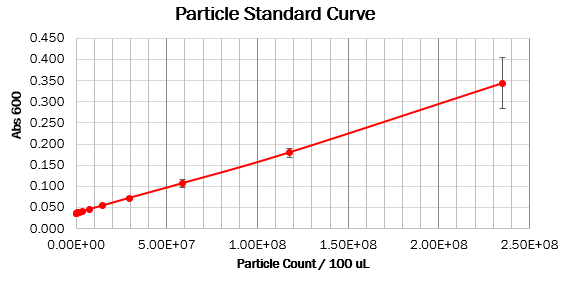
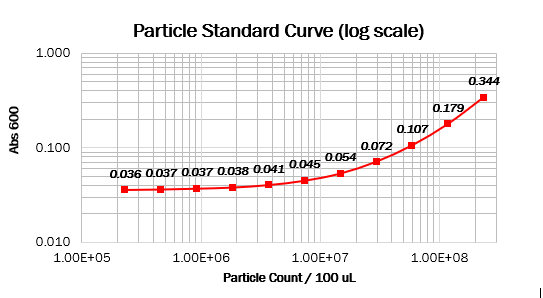
| + | === Calibration 2: particle standard curve === | ||
We prepared dilution series of monodisperse silica microspheres and measured the Abs600 in our plate reader. The size and optical characteristics of these microspheres are similar to cells, and there is a known amount of particles per volume. This measurement allowed us to construct a standard curve of particle concentration which can be used to convert Abs600 measurements to an estimated number of cells. | We prepared dilution series of monodisperse silica microspheres and measured the Abs600 in our plate reader. The size and optical characteristics of these microspheres are similar to cells, and there is a known amount of particles per volume. This measurement allowed us to construct a standard curve of particle concentration which can be used to convert Abs600 measurements to an estimated number of cells. | ||
| − | + | [[File:T--Aix-Marseille--G1.png|600px|center|]] | |
| + | [[File:T--Aix-Marseille--G2.png|600px|center|]] | ||
Values should form a straight line, with a 1:1 slope, on both linear and log scale curves. The particle standard curve forms a perfect line with a partial 1:1 slope. A possible explanation suggests a consistent pipetting error causing this partial 1:1 slope. Furthermore, the particle standard curve (log scale) doesn’t form a linear curve. To obtain the wanted curve, we should form a baseline for our measurements where we neglect the water’s absorbance at 600 nm. | Values should form a straight line, with a 1:1 slope, on both linear and log scale curves. The particle standard curve forms a perfect line with a partial 1:1 slope. A possible explanation suggests a consistent pipetting error causing this partial 1:1 slope. Furthermore, the particle standard curve (log scale) doesn’t form a linear curve. To obtain the wanted curve, we should form a baseline for our measurements where we neglect the water’s absorbance at 600 nm. | ||
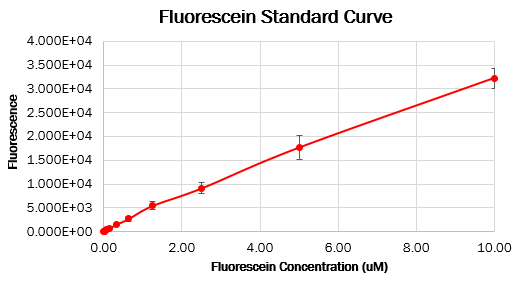
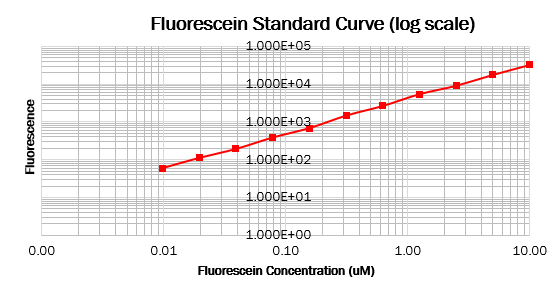
| − | === Calibration 3: | + | === Calibration 3: fluorescence standard curve === |
To make the standard curve of fluorescence for fluorescein concentration, we made a dilution series of fluorescein in a 96 wells black plate and measured fluorescence with the same settings as the cell measurements. With this standard curve, we could correct our cell measurements to an equivalent fluorescein concentration, ultimately converting this into a concentration of GFP. | To make the standard curve of fluorescence for fluorescein concentration, we made a dilution series of fluorescein in a 96 wells black plate and measured fluorescence with the same settings as the cell measurements. With this standard curve, we could correct our cell measurements to an equivalent fluorescein concentration, ultimately converting this into a concentration of GFP. | ||
| − | + | [[File:T--Aix-Marseille--G3.png|600px|center|]] | |
| + | [[File:T--Aix-Marseille--G4.png|600px|center|]] | ||
=== Cell measurements === | === Cell measurements === | ||
| − | After obtaining OD600 and fluorescence raw readings, the fluorescence per OD ratio was calculated to determine each promotor’s strength. We see that the promoter ([ | + | After obtaining OD600 and fluorescence raw readings, the fluorescence per OD ratio was calculated to determine each promotor’s strength. We see that the promoter ([http://parts.igem.org/Part:BBa_J23101 J23101]) in test device 1 ([http://parts.igem.org/Part:BBa_J364000 BBa_J364000]) and the promoter ([http://parts.igem.org/Part:BBa_J23100 J23100]) in test device 4 ([http://parts.igem.org/Part:BBa_J364007 BBa_J364007]) are indeed the strongest promoters, as the GFP expression seems high. However, the promoter ([http://parts.igem.org/Part:BBa_J23106 J23106]) in test device 2 ([http://parts.igem.org/Part:BBa_J364001 BBa_J364001]) and the promoter ([http://parts.igem.org/Part:BBa_J23116 J23116]) in device 6 ([http://parts.igem.org/Part:BBa_J364009 BBa_J364009]) has a medium strength, as the GFP expression was observed to have a medium increase. Whereas the promoter ([http://parts.igem.org/Part:BBa_J23117 J23117]) in test device 3 ([http://parts.igem.org/Part:BBa_J364002 BBa_J364002]) and the promoter ([http://parts.igem.org/Part:BBa_J23104 J23104]) in test device 5 ([http://parts.igem.org/Part:BBa_J364008 BBa_J364008]) showed have a low strength, as the GFP expression showed a low increase. |
| + | |||
| + | [[File:T--Aix-Marseille--G5.png |600px|center|]] | ||
| + | |||
| + | These results suggest that the promoter J23101 and J23100 had the strongest affinity for the RNA polymerase when compared with other promoters. Almost all ''E. coli'' strains show a high OD600 value, suggesting a relatively unaffected growth. Thus, the transformed plasmids aren’t toxic to the cells. The table below shows the measured strength of the Anderson promoter collection as displayed in the iGEM registry of standard biological parts. The experimental findings are compatible. | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |||
| + | ! style="width: 15%;;" | Promoter | ||
| + | ! style="width: 15%;" | Measured strength | ||
| + | |- | ||
| + | | BBa_J23100 | ||
| + | | 1 | ||
| + | |- | ||
| + | | BBa_J23101 | ||
| + | | 0.70 | ||
| + | |- | ||
| + | | BBa_J23104 | ||
| + | | 0.72 | ||
| + | |- | ||
| + | | BBa_J23106 | ||
| + | | 0.47 | ||
| + | |- | ||
| + | | BBa_J23116 | ||
| + | | 0.16 | ||
| + | |- | ||
| + | | BBa_J23117 | ||
| + | | 0.06 | ||
| + | |} | ||
| + | |||
| + | === CFUs (Colony Forming Units) === | ||
| + | |||
| + | This procedure can be used to calibrate OD600 to colony forming unit (CFUs) counts, which are directly relatable to the cell concentration of the culture (i.e. viable cell counts per mL). This protocol assumes that 1 bacterial cell will give rise to 1 colony. For the CFU protocol, we counted colonies for our two positive control ([http://parts.igem.org/Part:BBa_I20270 BBa_I20270]) cultures and our two negative control ([http://parts.igem.org/Part:BBa_R0040 BBa_R0040]) cultures. cultures and our two negative control cultures. | ||
| + | |||
| + | We made overnight cultures. The following day, the OD600 values were measured and diluted to a target OD600 of 0.1. Afterward, we made dilution series (5 dilutions with a 1/10 dilution factor each) with our starting samples. Dilutions 3, 4 and 5 were incubated overnight on LB + agar plates. | ||
| + | |||
| + | Based on the assumption that 1 bacterial cell gives rise to 1 colony, colony forming units (CFUs) per 1mL of an OD600 = 0.1 culture can be calculated following two steps: counting the colonies on each plate with fewer than 300 colonies, then multiplying the colony count by the final dilution factor on each plate. | ||
| + | |||
| + | For the 8 x 10^4 dilution factor plates, all of them had too many colonies to count (over 400 colonies). This is considered "too numerous to count" (TNTC). Whereas, for the 8 x 10^5 dilution and the 8 x 10^6 factor plates, all of them had countable colonies. | ||
| − | + | {| class="wikitable" | |
| + | ! style="width: 15%;;" | Plate | ||
| + | ! style="width: 15%;" | 8 x 10^4 dilution | ||
| + | ! style="width: 15%;" | 8 x 10^5 dilution | ||
| + | ! style="width: 15%;" | 8 x 10^6 dilution | ||
| + | ! style="width: 15%;" | CFU/mL | ||
| + | |- | ||
| + | | 1.1 | ||
| + | | TNTC | ||
| + | | 440 | ||
| + | | 59 | ||
| + | | 4.72 x 10^8 | ||
| + | |- | ||
| + | | 1.2 | ||
| + | | TNTC | ||
| + | | 430 | ||
| + | | 49 | ||
| + | | 3.92 x 10^8 | ||
| + | |- | ||
| + | | 1.3 | ||
| + | | TNTC | ||
| + | | 413 | ||
| + | | 44 | ||
| + | | 3.52 x 10^8 | ||
| + | |- | ||
| + | | 2.1 | ||
| + | | TNTC | ||
| + | | 400 | ||
| + | | 46 | ||
| + | | 3.68 x 10^8 | ||
| + | |- | ||
| + | | 2.2 | ||
| + | | TNTC | ||
| + | | 324 | ||
| + | | 40 | ||
| + | | 3.2 x 10^8 | ||
| + | |- | ||
| + | | 2.3 | ||
| + | | TNTC | ||
| + | | 388 | ||
| + | | 52 | ||
| + | | 4.16 x 10^8 | ||
| + | |- | ||
| + | | 3.1 | ||
| + | | TNTC | ||
| + | | 433 | ||
| + | | 41 | ||
| + | | 3.28 x 10^8 | ||
| + | |- | ||
| + | | 3.2 | ||
| + | | TNTC | ||
| + | | 422 | ||
| + | | 44 | ||
| + | | 3.52 x 10^8 | ||
| + | |- | ||
| + | | 3.3 | ||
| + | | TNTC | ||
| + | | 294 | ||
| + | | 55 | ||
| + | | 4.4 x 10^8 | ||
| + | |- | ||
| + | | 4.1 | ||
| + | | TNTC | ||
| + | | 241 | ||
| + | | 21 | ||
| + | | 4.72 x 10^8 | ||
| + | |- | ||
| + | | 4.2 | ||
| + | | TNTC | ||
| + | | 362 | ||
| + | | 30 | ||
| + | | 1.68 x 10^8 | ||
| + | |- | ||
| + | | 4.3 | ||
| + | | TNTC | ||
| + | | 289 | ||
| + | | 32 | ||
| + | | 2.56 x 10^8 | ||
| + | |- | ||
| + | |} | ||
| − | + | Almost all ''E. coli'' strains, with the positive and negative controls plasmids, show an identical CFU, suggesting a relatively unaffected growth. Thus, high expression of the GFP protein isn’t toxic to the cells. | |
Latest revision as of 23:15, 17 October 2018
Interlab
Brief history
Over the past years, iGEM has advanced the frontiers of science with the biggest interlaboratory study ever done in synthetic biology. These studies established a baseline for replicability of fluorescence measurements and identified likely key sources of error and have now been published as an open-access journal article in PLOS ONE.
The InterLab study has been developing a robust measurement procedure for green fluorescent protein (GFP) over the last several years. GFP was chosen as the measurement marker for this study since it's one of the most used markers in synthetic biology and, as a result, most laboratories are equipped to measure this protein.
The fifth interlab study
The fifth international InterLab study aims to identify and correct the sources of systematic variability in synthetic biology measurements so that eventually, measurements that are taken in different labs will be no more variable than measurements taken within the same lab. This year, we helped in answering the following question: Can we reduce lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs) instead of OD?
Materials and methods
Cell measurments
For this study, we transformed six different plasmids, along with a negative and positive control into E. coli K-12 DH5-alpha. The plasmids, from here on referred to as devices, contain a constitutive promoter with low, medium or high strengths. The test devices express GFP (as a reporter in the pSB1C3 backbone). A plasmid containing a constitutive promoter and GFP sequence (BBa_I20270) and a plasmid containing a TetR repressible promoter (BBa_R0040) in the pSB1C3 backbone, were used as positive and negative controls. All plasmids were delivered to us in the iGEM 2018 distribution kit.
We made overnight cultures of two colonies of each transformation plate. The following day, the OD600 values of the overnight cultures were measured, diluted to a target OD600 of 0.02 and grown over 6 hours. Afterward, the absorbance (600 nm) and fluorescence (ex. 485 nm – em. 520 nm, gain = 50) of the growing cultures were measured in a 96 wells black plate in a plate reader (TECAN M200 Infinite) at 0, 3 and 6 hours.
Colony forming units (CFUs)
This procedure can be used to calibrate OD600 to colony forming unit (CFUs) counts, which are directly relatable to the cell concentration of the culture (i.e. viable cell counts per mL). This protocol assumes that 1 bacterial cell will give rise to 1 colony. For the CFU protocol, we counted colonies for our two positive control (BBa_I20270) cultures and our two negative control (BBa_R0040) cultures.
We made overnight cultures. The following day, the OD600 values were measured and diluted to a target OD600 of 0.1. Afterward, we made a dilution series (5 dilutions) with our starting samples. Dilutions 3, 4 and 5 were incubated overnight on LB + agar plates.
Based on the assumption that 1 bacterial cell gives rise to 1 colony, colony forming units (CFU) per 1mL of an OD600 = 0.1 culture can be calculated following two steps: counting the colonies on each plate with fewer than 300 colonies, then multiplying the colony count by the final dilution factor on each plate.
Results
Calibration 1: reference point
We measured Abs600 of LUDOX CL-X and H2O to use them as a reference point to obtain a ratiometric conversion factor. By multiplying this factor with the raw Abs600 measurement data, we could transform our absorbance data into a standard OD600 measurement, accounting for instrument differences.
| LUDOX CL-X | H2O | |
|---|---|---|
| Replicate 1 | 0.0537 | 0.036 |
| Replicate 2 | 0.054099999 | 0.0359 |
| Replicate 3 | 0.056299999 | 0.0361 |
| Replicate 4 | 0.056499999 | 0.0361 |
| Arithmetic Mean | 0.055 | 0.036 |
| Corrected Abs600 | 0.019 | |
| Reference OD600 | 0.063 | |
| OD600/Abs600 | 3.294 |
Calibration 2: particle standard curve
We prepared dilution series of monodisperse silica microspheres and measured the Abs600 in our plate reader. The size and optical characteristics of these microspheres are similar to cells, and there is a known amount of particles per volume. This measurement allowed us to construct a standard curve of particle concentration which can be used to convert Abs600 measurements to an estimated number of cells.
Values should form a straight line, with a 1:1 slope, on both linear and log scale curves. The particle standard curve forms a perfect line with a partial 1:1 slope. A possible explanation suggests a consistent pipetting error causing this partial 1:1 slope. Furthermore, the particle standard curve (log scale) doesn’t form a linear curve. To obtain the wanted curve, we should form a baseline for our measurements where we neglect the water’s absorbance at 600 nm.
Calibration 3: fluorescence standard curve
To make the standard curve of fluorescence for fluorescein concentration, we made a dilution series of fluorescein in a 96 wells black plate and measured fluorescence with the same settings as the cell measurements. With this standard curve, we could correct our cell measurements to an equivalent fluorescein concentration, ultimately converting this into a concentration of GFP.
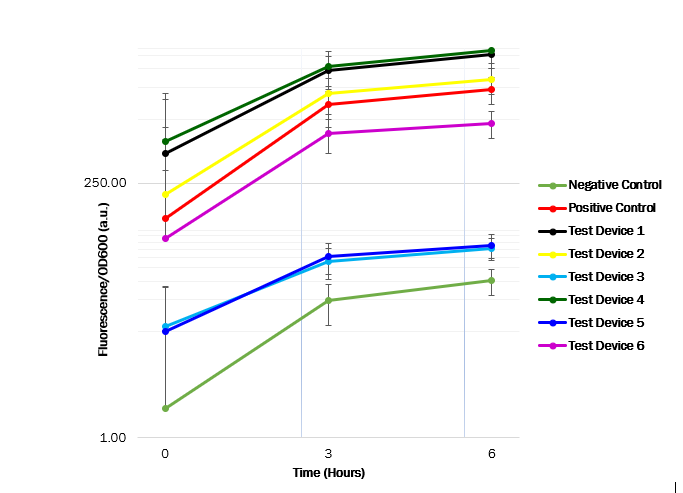
Cell measurements
After obtaining OD600 and fluorescence raw readings, the fluorescence per OD ratio was calculated to determine each promotor’s strength. We see that the promoter (J23101) in test device 1 (BBa_J364000) and the promoter (J23100) in test device 4 (BBa_J364007) are indeed the strongest promoters, as the GFP expression seems high. However, the promoter (J23106) in test device 2 (BBa_J364001) and the promoter (J23116) in device 6 (BBa_J364009) has a medium strength, as the GFP expression was observed to have a medium increase. Whereas the promoter (J23117) in test device 3 (BBa_J364002) and the promoter (J23104) in test device 5 (BBa_J364008) showed have a low strength, as the GFP expression showed a low increase.
These results suggest that the promoter J23101 and J23100 had the strongest affinity for the RNA polymerase when compared with other promoters. Almost all E. coli strains show a high OD600 value, suggesting a relatively unaffected growth. Thus, the transformed plasmids aren’t toxic to the cells. The table below shows the measured strength of the Anderson promoter collection as displayed in the iGEM registry of standard biological parts. The experimental findings are compatible.
| Promoter | Measured strength |
|---|---|
| BBa_J23100 | 1 |
| BBa_J23101 | 0.70 |
| BBa_J23104 | 0.72 |
| BBa_J23106 | 0.47 |
| BBa_J23116 | 0.16 |
| BBa_J23117 | 0.06 |
CFUs (Colony Forming Units)
This procedure can be used to calibrate OD600 to colony forming unit (CFUs) counts, which are directly relatable to the cell concentration of the culture (i.e. viable cell counts per mL). This protocol assumes that 1 bacterial cell will give rise to 1 colony. For the CFU protocol, we counted colonies for our two positive control (BBa_I20270) cultures and our two negative control (BBa_R0040) cultures. cultures and our two negative control cultures.
We made overnight cultures. The following day, the OD600 values were measured and diluted to a target OD600 of 0.1. Afterward, we made dilution series (5 dilutions with a 1/10 dilution factor each) with our starting samples. Dilutions 3, 4 and 5 were incubated overnight on LB + agar plates.
Based on the assumption that 1 bacterial cell gives rise to 1 colony, colony forming units (CFUs) per 1mL of an OD600 = 0.1 culture can be calculated following two steps: counting the colonies on each plate with fewer than 300 colonies, then multiplying the colony count by the final dilution factor on each plate.
For the 8 x 10^4 dilution factor plates, all of them had too many colonies to count (over 400 colonies). This is considered "too numerous to count" (TNTC). Whereas, for the 8 x 10^5 dilution and the 8 x 10^6 factor plates, all of them had countable colonies.
| Plate | 8 x 10^4 dilution | 8 x 10^5 dilution | 8 x 10^6 dilution | CFU/mL |
|---|---|---|---|---|
| 1.1 | TNTC | 440 | 59 | 4.72 x 10^8 |
| 1.2 | TNTC | 430 | 49 | 3.92 x 10^8 |
| 1.3 | TNTC | 413 | 44 | 3.52 x 10^8 |
| 2.1 | TNTC | 400 | 46 | 3.68 x 10^8 |
| 2.2 | TNTC | 324 | 40 | 3.2 x 10^8 |
| 2.3 | TNTC | 388 | 52 | 4.16 x 10^8 |
| 3.1 | TNTC | 433 | 41 | 3.28 x 10^8 |
| 3.2 | TNTC | 422 | 44 | 3.52 x 10^8 |
| 3.3 | TNTC | 294 | 55 | 4.4 x 10^8 |
| 4.1 | TNTC | 241 | 21 | 4.72 x 10^8 |
| 4.2 | TNTC | 362 | 30 | 1.68 x 10^8 |
| 4.3 | TNTC | 289 | 32 | 2.56 x 10^8 |
Almost all E. coli strains, with the positive and negative controls plasmids, show an identical CFU, suggesting a relatively unaffected growth. Thus, high expression of the GFP protein isn’t toxic to the cells.