(→Human pratices) |
|||
| (69 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{Aix-Marseille}} | + | {{Aix-Marseille|title=Results}} __NOTOC__ |
| − | + | ||
| + | Positive thoughts, positive actions and positive results. As a hard working team, we aimed high this year. We have generated results that met with the bronze, silver and gold medals requiremements. We were able to produce a functional endochitinase (Gold medal criterion), metabolic intermediates from the benzaldehyde/benzylalcohol biosynthesis pathway, and the DMTS/DMDS pheromones (Silver medal criterion) . We have successfully modeled the trap (Gold medal criterion) and tested it in infested premises. Furthermore, we have made collaborations (Silver medal criterion) with the iGEM committee, as well as with private companies. This sets a solid ground for further developments around the project. Last but not least, a nationwide advertisement campaign engaged the public in our adventure. We had a mutual interest: all bed bugs must come to an end. | ||
| − | + | =Wetlab= | |
| − | + | ==[https://2018.igem.org/Team:Aix-Marseille/Experiments Endochitinase]== | |
| − | + | [[File:T--Aix-Marseille--(fourmis).jpg|250 px|right]] | |
| − | + | The endochitinase is a protein naturally produced by ''Beauveria bassiana'' an enthomopathogenic fungus. We wanted to overproduce it (in ''E. coli'') and extract it to enhance the killing efficieny and speed of the fungus propagation. | |
| + | We succesfully produced, and purified the functional endochitinase ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K2718022 BBa_K2718022]). | ||
| + | This part is an improvement of the part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1913000 BBa_K1913000] '''Gold medal criteria'''. | ||
| − | + | ==Pheromone production== | |
| − | + | ||
| − | + | ||
| + | The aim was to produce two types of pheromone: DMTS/DMDS and benzaldehyde/benzylalcohol. We did some research on the two molecules to get the biosynthesis pathways to produce them in ''E. coli''. First, we needed to add the three enzymes implicated in the biosynthesis of benzaldehyde/benzylalcohol: MdlB, MdlC and Hmas. We successfully cloned the Hmas gene ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K2718011 BBa_K2718011]) and produced the equivalent protein. This enabled [https://2018.igem.org/Team:Aix-Marseille/Experiments the production of the first metabolic intermediate] (S-Mandelate) of the pathway. | ||
| + | Second, we were able to clone the methionine-γ-lyase gene ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K2718006 BBa_K2718006]) and produce the equivalent protein. This enabled the achievement of a first goal: the production of the DMTS/DMDS pheromone. This part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K2718006 BBa_K2718006] is our validated part '''Silver medal criteria''' | ||
| + | ==Trap tests== | ||
| + | A few weeks before the giant jamboree we have decided to go all the way with the breaking bugs project. We took a preliminary [https://2018.igem.org/Team:Aix-Marseille/Design trap design] to the field to test the efficiency of both units: ''Beauveria bassiana'' combined with the adjuvants, and the pheromones efficiency. In collaboration with local infested residences, we had the chance to demonstrate some functional aspects about the prototype. | ||
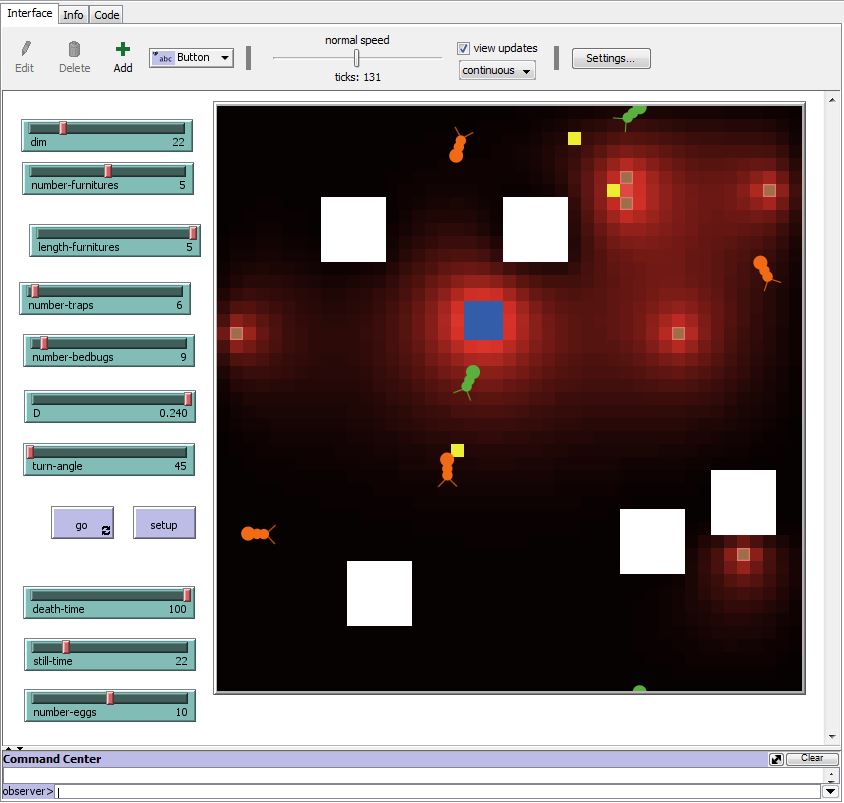
| + | ==[https://2018.igem.org/Team:Aix-Marseille/Model Model]== | ||
| + | [[File:T--Aix-Marseille--run bedbug 1.png|350 px|right]] | ||
| + | We decided to model the deployment of our bed bug trap in a realistic environment, to understand how it was likely to work and what parameters might be important. This task involved a number of different modeling tasks, that posed different problems: modelling the diffusion of pheromones in the room coming both from a natural bed bug nest and the traps that we plan to deploy; modelling the movement of bed bugs influenced both by the pheromone field and their nest; and modeling the fungal epidemic that we plan to induce in the bed bug population. Though the model is complex and includes many parameters, it has already allowed us to draw several conclusions, and as the model is improved and the parameters are refined other conclusions will follow. | ||
| + | For instance, using realistic diffusion parameters it is clear that the pheromone field is relatively rapidly established and so the precise nature and concentration of pheromones is probably not critical. In contrast, the delay between infection and death of bed bugs is critical for ensuring the eradication of the nest. Our modeling thus helps understand critical aspects of the proposed design. This is a '''Gold medal criteria'''. | ||
| − | + | ==[https://2018.igem.org/Team:Aix-Marseille/Interlab Interlab]== | |
| + | The InterLab study has been developing a robust measurement procedure for green fluorescent protein (GFP) over the last several years. GFP was chosen as the measurement marker for this study since it's one of the most used markers in synthetic biology and, as a result, most laboratories are equipped to measure this protein. This year, we helped in answering the following question: Can we reduce lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs) instead of OD? | ||
| + | We participated in this '''Bronze medal criteria''' | ||
| − | + | ==[https://2018.igem.org/Team:Aix-Marseille/Perspectives Perspectives]== | |
| − | + | The nationwide success of our approach to treat bed bugs has encouraged us to elaborate perspectives for long term development. We are collaborating with multiple potential investors we have met at the [https://2018.igem.org/Team:Aix-Marseille/SHUC social housing union convention] and [https://contact.rentokil.fr/fr-deratisation-v3/?utm_term=rentokil&gclid=CjwKCAjwu5veBRBBEiwAFTqDwYY8qKmbHDOWKutmcTcQWmW9qgyysHT0DWQj_fK3BRepTRwrgH8MIBoCjJQQAvD_BwE Rentokil] for further research and development around our plan. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ==[https://2018.igem.org/Team:Aix-Marseille/Collaborations Collaborations]== | |
| − | + | Connecting with other iGEM teams from all around the world was the power engine of our project. We managed to make collaborations on many levels. We helped the Mexican team with their project's advancement, the German team with their awareness campaign and the Chinese team with their international language project. Additionnally, we organized the 2nd edition of the Mediterranean meetup to help the fellow iGEMers prepare for the giant jamboree. This is a '''Silver Medal criteria''' | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | ==[https://2018.igem.org/Team:Aix-Marseille/Human_Practices Human Practices]== | ||
| − | + | The public engagement was the pillar of the breaking bugs project. After a succesfull nationwide adverstisement campaign, we were able to engage the french and international commitees in our plan. We had a solid validation by a massive waves of donations for our crowfunding campaign. | |
| − | + | Our human practices and public engagement was very strong, completing a '''Silver medal criteria''', and we integrated our human practices work into the project design a '''Gold medal criteria'''. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 02:00, 18 October 2018
Results
Positive thoughts, positive actions and positive results. As a hard working team, we aimed high this year. We have generated results that met with the bronze, silver and gold medals requiremements. We were able to produce a functional endochitinase (Gold medal criterion), metabolic intermediates from the benzaldehyde/benzylalcohol biosynthesis pathway, and the DMTS/DMDS pheromones (Silver medal criterion) . We have successfully modeled the trap (Gold medal criterion) and tested it in infested premises. Furthermore, we have made collaborations (Silver medal criterion) with the iGEM committee, as well as with private companies. This sets a solid ground for further developments around the project. Last but not least, a nationwide advertisement campaign engaged the public in our adventure. We had a mutual interest: all bed bugs must come to an end.
Wetlab
Endochitinase
The endochitinase is a protein naturally produced by Beauveria bassiana an enthomopathogenic fungus. We wanted to overproduce it (in E. coli) and extract it to enhance the killing efficieny and speed of the fungus propagation. We succesfully produced, and purified the functional endochitinase (BBa_K2718022). This part is an improvement of the part BBa_K1913000 Gold medal criteria.
Pheromone production
The aim was to produce two types of pheromone: DMTS/DMDS and benzaldehyde/benzylalcohol. We did some research on the two molecules to get the biosynthesis pathways to produce them in E. coli. First, we needed to add the three enzymes implicated in the biosynthesis of benzaldehyde/benzylalcohol: MdlB, MdlC and Hmas. We successfully cloned the Hmas gene (BBa_K2718011) and produced the equivalent protein. This enabled the production of the first metabolic intermediate (S-Mandelate) of the pathway. Second, we were able to clone the methionine-γ-lyase gene (BBa_K2718006) and produce the equivalent protein. This enabled the achievement of a first goal: the production of the DMTS/DMDS pheromone. This part BBa_K2718006 is our validated part Silver medal criteria
Trap tests
A few weeks before the giant jamboree we have decided to go all the way with the breaking bugs project. We took a preliminary trap design to the field to test the efficiency of both units: Beauveria bassiana combined with the adjuvants, and the pheromones efficiency. In collaboration with local infested residences, we had the chance to demonstrate some functional aspects about the prototype.
Model
We decided to model the deployment of our bed bug trap in a realistic environment, to understand how it was likely to work and what parameters might be important. This task involved a number of different modeling tasks, that posed different problems: modelling the diffusion of pheromones in the room coming both from a natural bed bug nest and the traps that we plan to deploy; modelling the movement of bed bugs influenced both by the pheromone field and their nest; and modeling the fungal epidemic that we plan to induce in the bed bug population. Though the model is complex and includes many parameters, it has already allowed us to draw several conclusions, and as the model is improved and the parameters are refined other conclusions will follow.
For instance, using realistic diffusion parameters it is clear that the pheromone field is relatively rapidly established and so the precise nature and concentration of pheromones is probably not critical. In contrast, the delay between infection and death of bed bugs is critical for ensuring the eradication of the nest. Our modeling thus helps understand critical aspects of the proposed design. This is a Gold medal criteria.
Interlab
The InterLab study has been developing a robust measurement procedure for green fluorescent protein (GFP) over the last several years. GFP was chosen as the measurement marker for this study since it's one of the most used markers in synthetic biology and, as a result, most laboratories are equipped to measure this protein. This year, we helped in answering the following question: Can we reduce lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs) instead of OD? We participated in this Bronze medal criteria
Perspectives
The nationwide success of our approach to treat bed bugs has encouraged us to elaborate perspectives for long term development. We are collaborating with multiple potential investors we have met at the social housing union convention and Rentokil for further research and development around our plan.
Collaborations
Connecting with other iGEM teams from all around the world was the power engine of our project. We managed to make collaborations on many levels. We helped the Mexican team with their project's advancement, the German team with their awareness campaign and the Chinese team with their international language project. Additionnally, we organized the 2nd edition of the Mediterranean meetup to help the fellow iGEMers prepare for the giant jamboree. This is a Silver Medal criteria
Human Practices
The public engagement was the pillar of the breaking bugs project. After a succesfull nationwide adverstisement campaign, we were able to engage the french and international commitees in our plan. We had a solid validation by a massive waves of donations for our crowfunding campaign. Our human practices and public engagement was very strong, completing a Silver medal criteria, and we integrated our human practices work into the project design a Gold medal criteria.