| Line 88: | Line 88: | ||
| − | == | + | === Chitinase production:=== |
| + | We achieve to produce chitinase thanks to our construction. A inducible surproduction by pLac promoter was used to study chitinase production, induction or none-induction experiments by IPTG can show the presence of the protein. We choose to produce chitinase by E. coli K-12 DH5-alpha strain and by BL21 strain. | ||
| + | For this two strains two differents recombinated plasmids with chit1 were tested, plasmid N°10 and N°11. | ||
| + | 1) DH5alpha production : | ||
| − | + | [[File:T--Aix-Marseille--ChitinaseResults1.jpg|600px|center|]] | |
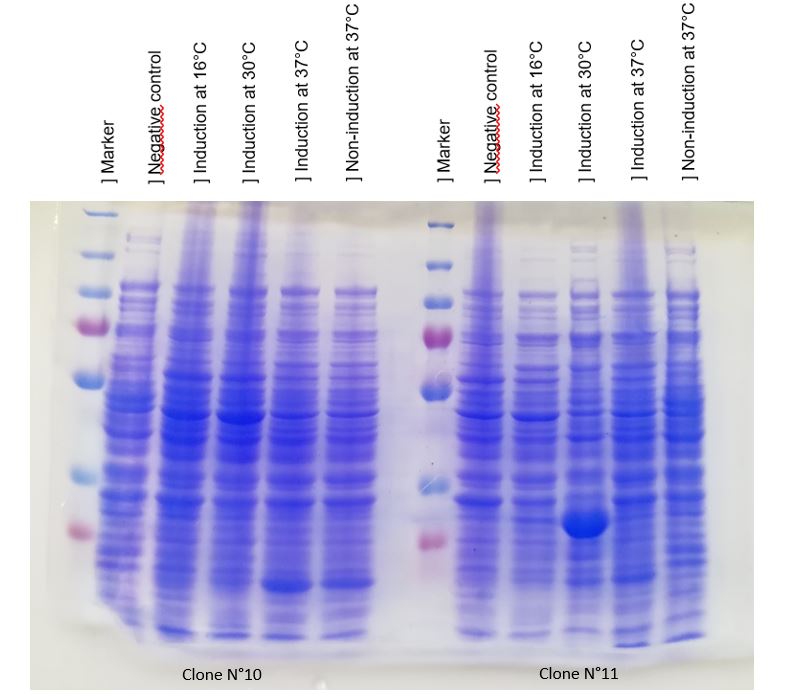
| − | + | Figure 1: SDS-PAGE of chitinase produced by E. coli K-12 DH5-alpha. | |
| − | + | As seen in Figure 1, a protein at 30 kDa was surproduced only at 30°C in DH5alpha. The non-induction by IPTG show an disappearance of the protein surproduced. We can concluded that chitinase is surproduced only at 30°C by DH5alpha strain, but it isn't normal that clone N°11 wasn't able to produced at least a low chitinase quantity. we will clarify this, thanks to a western blot. | |
| + | |||
| + | |||
| + | We can conclude that chitinase was only obtained thanks to plasmid N°11 by two strains, that was revealed by the obtention of a large band surrounding 30 kDa ( lenght of chitinase chit1). | ||
Revision as of 19:56, 11 October 2018
{{{title}}}
Results
Here you can describe the results of your project and your future plans.
What should this page contain?
- Clearly and objectively describe the results of your work.
- Future plans for the project.
- Considerations for replicating the experiments.
Describe what your results mean
- Interpretation of the results obtained during your project. Don't just show a plot/figure/graph/other, tell us what you think the data means. This is an important part of your project that the judges will look for.
- Show data, but remember all measurement and characterization data must be on part pages in the Registry.
- Consider including an analysis summary section to discuss what your results mean. Judges like to read what you think your data means, beyond all the data you have acquired during your project.
Project Achievements
You can also include a list of bullet points (and links) of the successes and failures you have had over your summer. It is a quick reference page for the judges to see what you achieved during your summer.
- A list of linked bullet points of the successful results during your project
- A list of linked bullet points of the unsuccessful results during your project. This is about being scientifically honest. If you worked on an area for a long time with no success, tell us so we know where you put your effort.
Inspiration
See how other teams presented their results.
Results
Chitinase production:
We achieve to produce chitinase thanks to our construction. A inducible surproduction by pLac promoter was used to study chitinase production, induction or none-induction experiments by IPTG can show the presence of the protein. We choose to produce chitinase by E. coli K-12 DH5-alpha strain and by BL21 strain. For this two strains two differents recombinated plasmids with chit1 were tested, plasmid N°10 and N°11.
1) DH5alpha production :
Figure 1: SDS-PAGE of chitinase produced by E. coli K-12 DH5-alpha.
As seen in Figure 1, a protein at 30 kDa was surproduced only at 30°C in DH5alpha. The non-induction by IPTG show an disappearance of the protein surproduced. We can concluded that chitinase is surproduced only at 30°C by DH5alpha strain, but it isn't normal that clone N°11 wasn't able to produced at least a low chitinase quantity. we will clarify this, thanks to a western blot.
We can conclude that chitinase was only obtained thanks to plasmid N°11 by two strains, that was revealed by the obtention of a large band surrounding 30 kDa ( lenght of chitinase chit1).