ElianDupre (Talk | contribs) |
ElianDupre (Talk | contribs) |
||
| Line 96: | Line 96: | ||
<table style="width: 30%; float: right;" > | <table style="width: 30%; float: right;" > | ||
<tr><td><a href="https://static.igem.org/mediawiki/2018/c/c3/T--OLS_Canmore_Canada--PD.jpg" target="_blank"><img width="100%"src="https://static.igem.org/mediawiki/2018/c/c3/T--OLS_Canmore_Canada--PD.jpg"></a></td></tr> | <tr><td><a href="https://static.igem.org/mediawiki/2018/c/c3/T--OLS_Canmore_Canada--PD.jpg" target="_blank"><img width="100%"src="https://static.igem.org/mediawiki/2018/c/c3/T--OLS_Canmore_Canada--PD.jpg"></a></td></tr> | ||
| − | <tr><td class="imagecaptiontext"> | + | <tr><td class="imagecaptiontext">Figure-1</td></tr> |
</table> | </table> | ||
<p style="max-width: 60%;"> | <p style="max-width: 60%;"> | ||
When examining our project, we need to consider how long it will take for the proteins to secrete out of the chassis organism. This is where our basic models come into play. We used these models to determine approximately how much of the protein our bacteria would be able to produce over any given 24 hour period. We began this process by researching how long, on average, it may take our bacteria, B.subtilis, to produce and secrete this protein. With our results, we continued on further, creating a graph (Figure- 1) portraying this data.The first graph (Figure-1) was based off of the assumption that all of the protein created inside of the cell in a 24 hour period would then also be completely secreted by cell. This was done to get an average baseline, which might then be used later as a way to determine if the data graphed later on still followed the same general linear path so that we may be able to see approximations and identify clear outliers. | When examining our project, we need to consider how long it will take for the proteins to secrete out of the chassis organism. This is where our basic models come into play. We used these models to determine approximately how much of the protein our bacteria would be able to produce over any given 24 hour period. We began this process by researching how long, on average, it may take our bacteria, B.subtilis, to produce and secrete this protein. With our results, we continued on further, creating a graph (Figure- 1) portraying this data.The first graph (Figure-1) was based off of the assumption that all of the protein created inside of the cell in a 24 hour period would then also be completely secreted by cell. This was done to get an average baseline, which might then be used later as a way to determine if the data graphed later on still followed the same general linear path so that we may be able to see approximations and identify clear outliers. | ||
<br> | <br> | ||
| + | <table style="width: 30%; float: right;" > | ||
| + | <tr><td><a href="https://static.igem.org/mediawiki/2018/c/cd/T--OLS_Canmore_Canada--SD.jpg " target="_blank"><img width="100%"src="https://static.igem.org/mediawiki/2018/c/cd/T--OLS_Canmore_Canada--SD.jpg "></a></td></tr> | ||
| + | <tr><td class="imagecaptiontext">Figure-2</td></tr> | ||
| + | </table> | ||
This creates the graph that you see as Figure-1. That is obviously not the case, as no bacteria is able to secrete 100% of all its protein. The next graph is a graph based on the assumption that not all the protein will be secreted and allows for an approximation of about a 10e^3 for each hour mark, as seen in figure-2. After that the next logical step, of course, was to run the test multiple times. This will yield a more accurate result in hopes to get a more precise representation of protein yield. As you can see, we are able to generate a result which closely matches that of our control line above. However, it is clear that through our calculations, some major outliers were found and marked in a different colour than black in our figure-3 graph. | This creates the graph that you see as Figure-1. That is obviously not the case, as no bacteria is able to secrete 100% of all its protein. The next graph is a graph based on the assumption that not all the protein will be secreted and allows for an approximation of about a 10e^3 for each hour mark, as seen in figure-2. After that the next logical step, of course, was to run the test multiple times. This will yield a more accurate result in hopes to get a more precise representation of protein yield. As you can see, we are able to generate a result which closely matches that of our control line above. However, it is clear that through our calculations, some major outliers were found and marked in a different colour than black in our figure-3 graph. | ||
</p> | </p> | ||
| + | <table style="width: 30%; float: right;" > | ||
| + | <tr><td><a href="https://static.igem.org/mediawiki/2018/7/77/T--OLS_Canmore_Canada--SMT.jpg " target="_blank"><img width="100%"src="https://static.igem.org/mediawiki/2018/7/77/T--OLS_Canmore_Canada--SMT.jpg "></a></td></tr> | ||
| + | <tr><td class="imagecaptiontext">Figure-3</td></tr> | ||
| + | </table> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Revision as of 12:09, 17 October 2018
MODELLING
Modelling
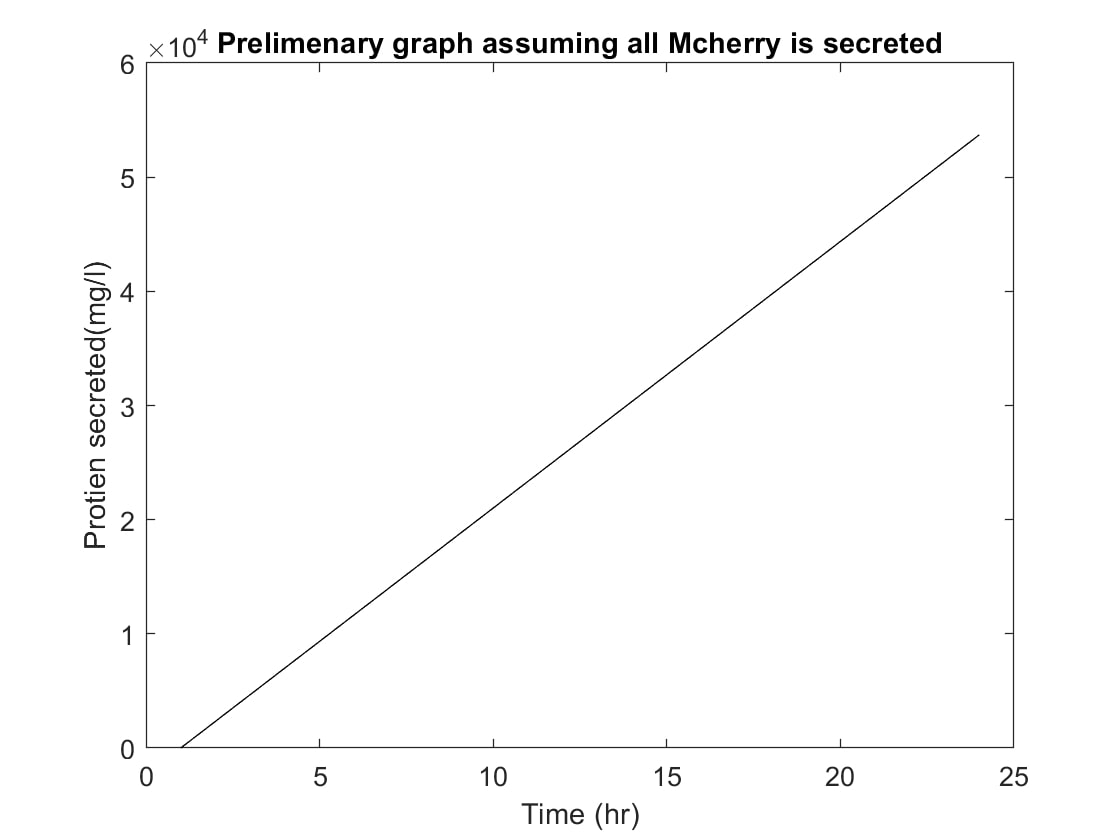 |
| Figure-1 |
When examining our project, we need to consider how long it will take for the proteins to secrete out of the chassis organism. This is where our basic models come into play. We used these models to determine approximately how much of the protein our bacteria would be able to produce over any given 24 hour period. We began this process by researching how long, on average, it may take our bacteria, B.subtilis, to produce and secrete this protein. With our results, we continued on further, creating a graph (Figure- 1) portraying this data.The first graph (Figure-1) was based off of the assumption that all of the protein created inside of the cell in a 24 hour period would then also be completely secreted by cell. This was done to get an average baseline, which might then be used later as a way to determine if the data graphed later on still followed the same general linear path so that we may be able to see approximations and identify clear outliers.
 |
| Figure-2 |
 |
| Figure-3 |

