(→Methionine gamma lyase production) |
(→Methionine gamma lyase production) |
||
| Line 65: | Line 65: | ||
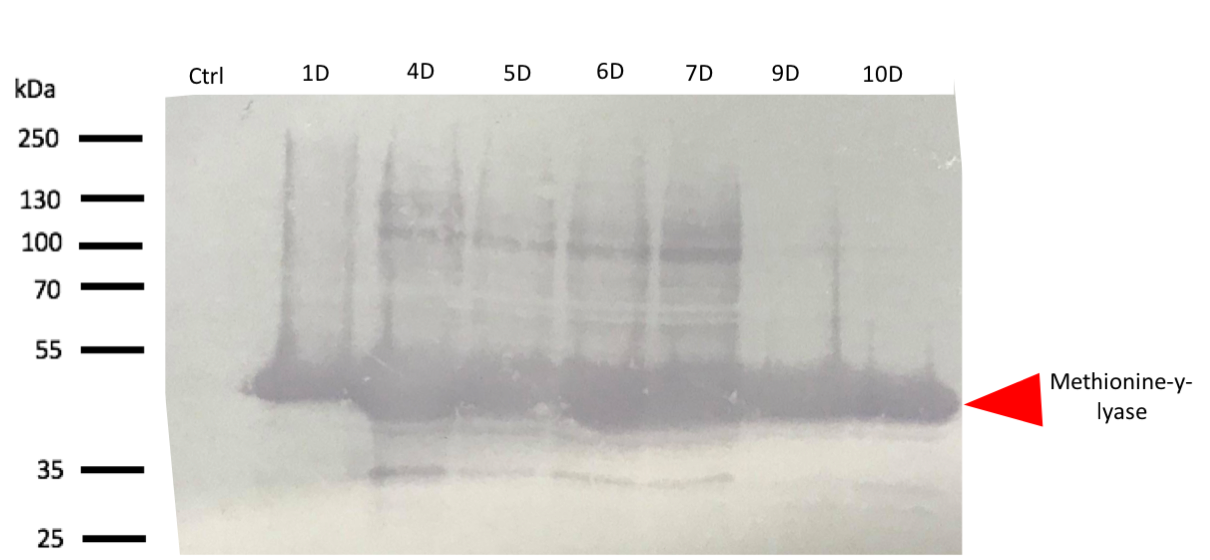
[[File:T--Aix-Marseille--WBDH5a.png|900px|center|thumb|Fig 1. SDS-PAGE DH5a, Empty plasmid (Ctrl), No-induced (1D), Induced at 30°C with 250mM IPTG(2D),Induced at 37°C with 500mM IPTG(3D), Induced at 37°C with 1000mM IPTG(4D),Induced at 30°C with 250mM IPTG(5D), Induced at 30°C with 500mM IPTG(6D), Induced at 30°C with 1000mM IPTG(7D), Induced at 16°C with 150mM IPTG(8D), Induced at 16°C with 500mM IPTG(9D), Induced at 16°C with 1000mM IPTG(10D)]] | [[File:T--Aix-Marseille--WBDH5a.png|900px|center|thumb|Fig 1. SDS-PAGE DH5a, Empty plasmid (Ctrl), No-induced (1D), Induced at 30°C with 250mM IPTG(2D),Induced at 37°C with 500mM IPTG(3D), Induced at 37°C with 1000mM IPTG(4D),Induced at 30°C with 250mM IPTG(5D), Induced at 30°C with 500mM IPTG(6D), Induced at 30°C with 1000mM IPTG(7D), Induced at 16°C with 150mM IPTG(8D), Induced at 16°C with 500mM IPTG(9D), Induced at 16°C with 1000mM IPTG(10D)]] | ||
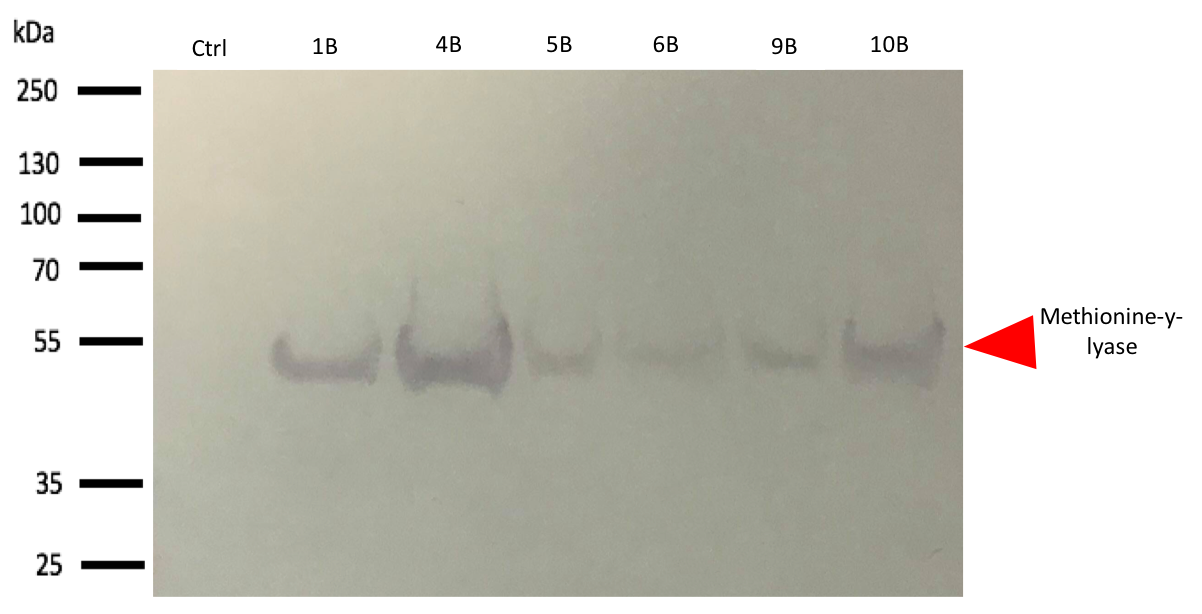
| − | The western blot thus confirmed the band identified on the SDS-PAGE corresponded to the methionine-γ-lyase produced by E.Coli DH5α bacteria. | + | [[File:T--Aix-Marseille--WBBL21.png--Aix-Marseille--WBBL21.png|900px|center|thumb|Fig 1. SDS-PAGE DH5a, Empty plasmid (Ctrl), No-induced (1B), Induced at 30°C with 250mM IPTG(2B),Induced at 37°C with 500mM IPTG(3B), Induced at 37°C with 1000mM IPTG(4B),Induced at 30°C with 250mM IPTG(5B), Induced at 30°C with 500mM IPTG(6B), Induced at 30°C with 1000mM IPTG(7B), Induced at 16°C with 150mM IPTG(8B), Induced at 16°C with 500mM IPTG(9B), Induced at 16°C with 1000mM IPTG(10B)]] |
| + | |||
| + | |||
| + | The western blot thus confirmed the band identified on the SDS-PAGE corresponded to the methionine-γ-lyase produced by E.Coli DH5α bacteria and by E.Coli BL21 bacteria. | ||
| + | |||
| + | For this test, it used 2 differents strains of E. coli Dh5 alpha which is similar to the strain DH1, and a BL21 strain. First, negatives controls are correct, there is no prodution of methionine-y-lyase with empty plasmid. Then, the bands to approximatively 47kDa, correspond to methionine-y-lyase molecular weight in both case . Moreover there does not seem to be any significant difference in protein production between the 4 samples when the temperature varies in the DH5 alpha case, whereas this parameter seems to influence production in BL21, since production seems less pronounced at 16 °C. Overall, the production of methionine-y-lyase is lower when the host is a BL21 strain compared to a production in a strain DH5 alpha. | ||
==Methionine gamma lyase purification== | ==Methionine gamma lyase purification== | ||
Revision as of 21:17, 17 October 2018
Experiments
Experiments
Describe the research, experiments, and protocols you used in your iGEM project. These should be detailed enough for another team to repeat your experiments.
What should this page contain?
- Protocols
- Experiments
- Documentation of the development of your project
Contents
Beauveria bassiana
We did some research on Beavearia bassiana fungus because it's an entomopathogenic fungus used during years in agriculture, especially in Canada. [1]
So, we wanted to use this fungus to fight bed bugs which invade our homes. We selected and bought a Beauveria sample.
Growing Beauveria bassiana
First of all, we grew Beauveria bassiana fungus on three different kind of culture medium :
- LB with an antibiotic Cm50
- Sabouraud with Cm 50
- Sabouraud with collagen and Cm 50 [https://2018.igem.org/Team:Aix-Marseille/Protocols]
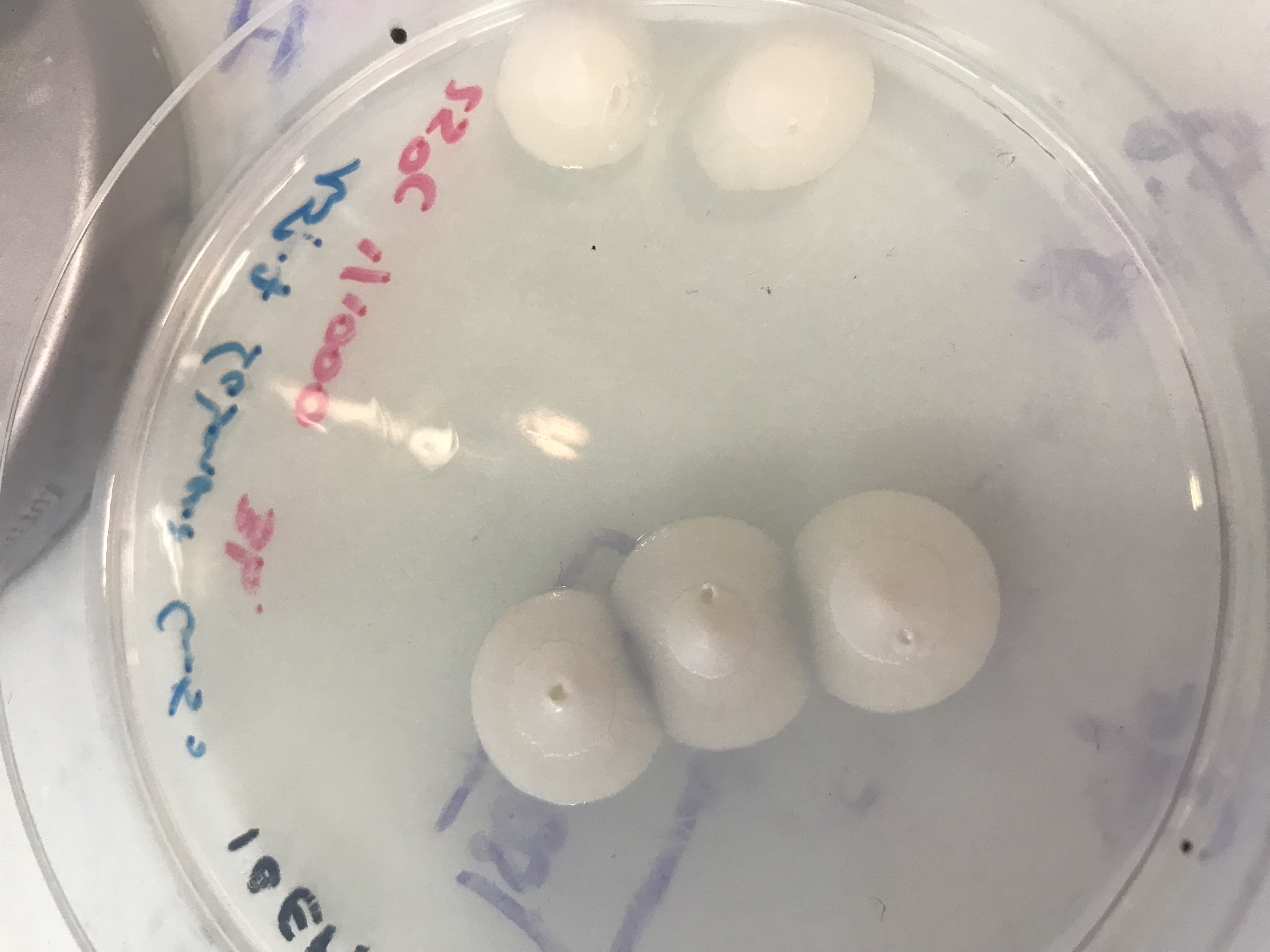
We made a serial dilution and let the fungus grow at three températures : 25°C, 30°C and 37°C. We obtained a lot of results but the culture medium and the most suitable temperature are the Sabouraud culture medium and 25°C. [2]
Killing insects with Beauveria bassiana
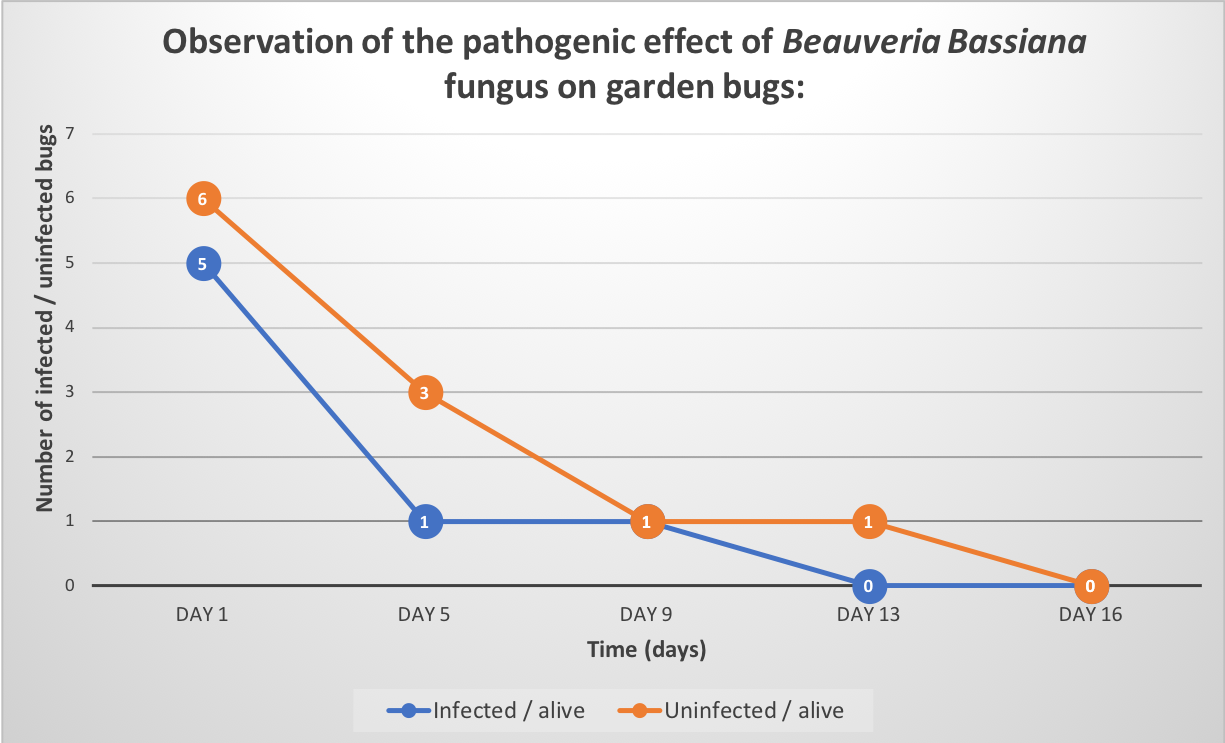
Secondly, we have tested Beauveria Sample on garden bugs with 4g of fungus sample diluted in 40ml of Tween 0,05% to confirmed that it was indeed Beauveria bassiana. We infected them by soaking and observed them for 2 weeks. Unfortunately, results obtained were inconclusive because we had problems keeping our control insect population alive.
However, we did observe mortality, and the infected bugs sprouted fungus a few days after the death of the insects. So, we decided to test our Beauveria Bassiana’s sample on bed bugs in real conditions.
Methionine gamma lyase
Methionine gamma lyase production
During these 3 months in the laboratory we managed to introduce a plasmid construct containing the LacI promoter inducible with IPTG (<bbpart>BBa_R0011</bbpart>), RBS + methionine-y-lyase (<bbpart>BBa_K1493300</bbpart>) and his-tag, inserted in a PSB1C3 vector. In order to test the production of the encoded protein, we did a SDS PAGE from several cellular supernatant as well as western blots. Regarding the his-tag, the latter has been added to methionine-y-lyase (<bbpart>BBa_K1493300</bbpart>) with the megaprimer PCR method. The part corresponding to methionine-y-lyase + his-tag is <bbpart>BBa_K2718005</bbpart>. The part that is used for the protein production is as follows <bbpart>BBa_K2718006</bbpart>.
To confirm the bands identified on SDS-PAGE corresponds to methionine-y-lyase, a western blot[3] with anti-his-tag antibodies was realized.
The western blot thus confirmed the band identified on the SDS-PAGE corresponded to the methionine-γ-lyase produced by E.Coli DH5α bacteria and by E.Coli BL21 bacteria.
For this test, it used 2 differents strains of E. coli Dh5 alpha which is similar to the strain DH1, and a BL21 strain. First, negatives controls are correct, there is no prodution of methionine-y-lyase with empty plasmid. Then, the bands to approximatively 47kDa, correspond to methionine-y-lyase molecular weight in both case . Moreover there does not seem to be any significant difference in protein production between the 4 samples when the temperature varies in the DH5 alpha case, whereas this parameter seems to influence production in BL21, since production seems less pronounced at 16 °C. Overall, the production of methionine-y-lyase is lower when the host is a BL21 strain compared to a production in a strain DH5 alpha.
Methionine gamma lyase purification
Methionine gamma lyase activity
Mandelate synthase
Mandelate synthase production
During these 3 months in the laboratory we managed to introduce a plasmid construct containing the IPTG promoter, the RBS and the hmaS gene, inserted in a PSB1C3 vector. In order to test the production of the encoded protein, we did a SDS PAGE from several cellular supernatant [4].
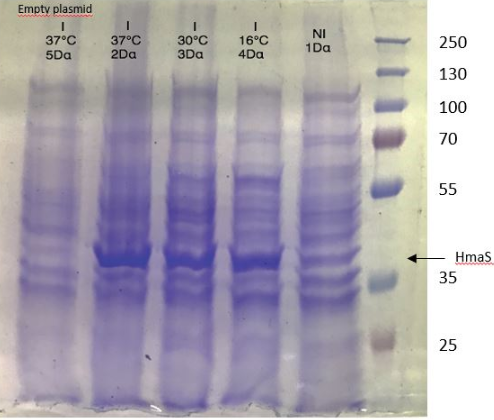
Figure 1 SDS-PAGE of HmaS production with induction (2, 3 and) 4, without induction ( 1,NI) and with empty plasmid (5), into a DH5 alpha E.coli strain.
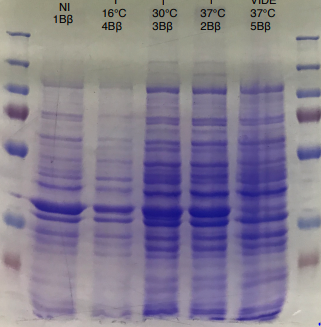
Figure 2 SDS-PAGE of HmaS production with induction (2, 3 and) 4, without induction ( 1,NI) and with empty plasmid (5), into a BL21 E.coli strain.
For this test we used 2 different strains of E. coli Dh5 alpha which is similar to the strain DH1 [5], and a BL21 strain [6].
First, negative control that we made are correct, there is no prodution of HmaS with empty plasmid. Then we can see bands to approximatively 37kDa, that correspond to HmaS molecular wheigt in both case [7]. Moreover there does not seem to be any significant difference in protein production between the 4 samples when the temperature varies in the DH5 alpha case, whereas this parameter seems to influence production in BL21, since production seems less pronounced at 16 °C. Overall, the production of HmaS appears to be lower when the host is a BL21 strain compared to a production in a strain DH5 alpha. However what we can say now is that we found what seems to be an HmaS overproduction.
Mandelate synthase purification
After doing the HmaS production tests and in order to obtain pure fractions of the protein, we subsequently purified the cell lysate of a strain of DH5 alpha transformed with our construct[8]. For this we did a purification on AKTA with an ion exchange column[9].
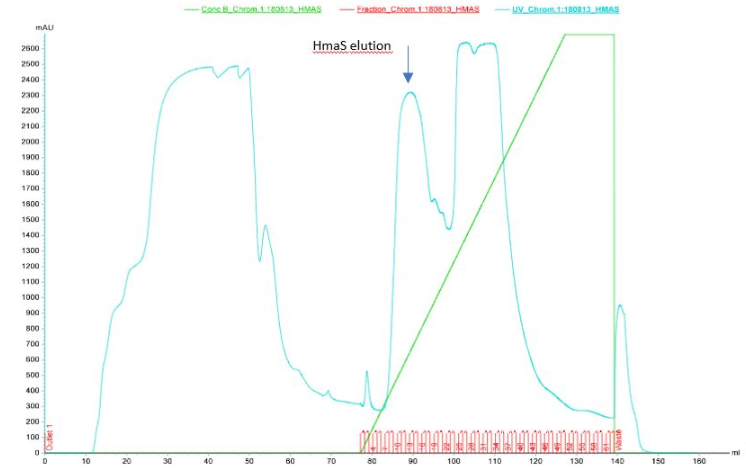
Figure 3 Elution of HmaS after purification by ion exchange column performed by Akta
After the first elution we can see 3 elution peaks in the graph which appear between 85 and 125 ml of solution B passage. One of them seems to be our protein however results are not perfect, because ion exchange chromatography is not the best technique to isolate protein. That is why we decided to do SDS-PAGE on the elution fractions to see if the protein is present (see figures 4a and 4b)[10]
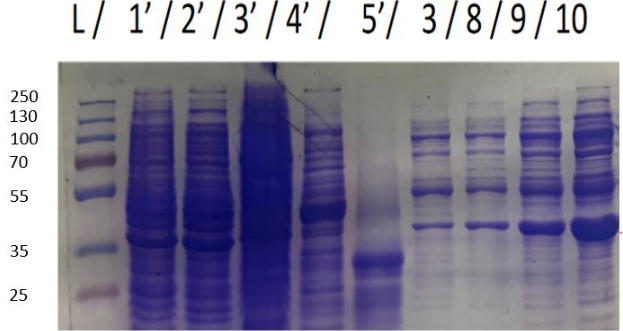
Figure 4a SDS-PAGE of eluate after purifiction L. Ladder , 1'Bacterial lysate ; 2'pellet of bacteria lysate ; 3' non purified fraction ; 4' breakthrought during protein injection ; 5' breakthrought during wash before elution ; 3,8,9 and 10 elution samples
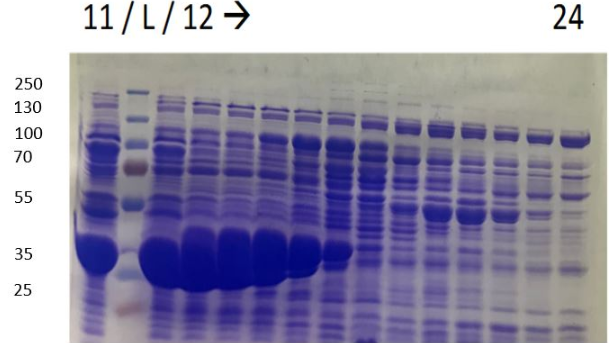
Figure 4b SDS-PAGE of eluate after purifiction L. Ladder 11 to 24 elution sample
Big spots, approxomatively to 37kDa, can be seen, which may correspond to HmaS. Nevertheless samples were not pure, others proteins were mixed with HmaS. However what we can say is that HmaS is on fractions 8 to 17, which correspond to the second peaks on figure 3. in conclusion, we partly purifed HmaS. Moreover, we can improve this purification, by using other methods like size eclusion chromatography.
Mandelate synthase activity
Then we tested the activity of our BBa_K2718011 biobrick [11]. In order to test the mandelate synthase activity in our E.coli strain, we have made an HPLC test on supernatent cells during 7h of cell culture as described in our protocols tab : [12]. The elution profils have been analysed and were compiled into several figures that can be found in the Notebook below.
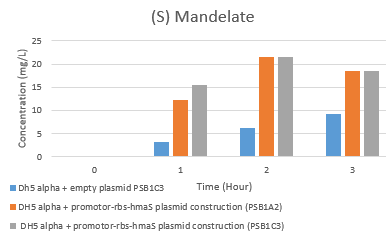
Fig 5e. Hplc test results: (S)-mandelate rate evolution
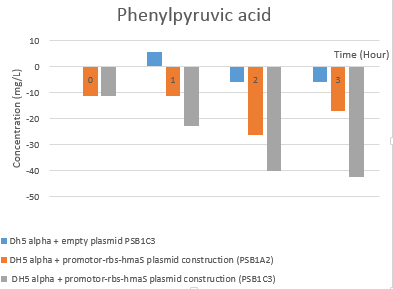
Fig 5f. Hplc test results: Phenylpyruvic acid rate evolution
On one hand the first thing that can be noticed is the global decrease and increase in phenylpyruvic acid and s mandelate concentrations, respectively, and whatever the tested condition, which might suggest that there is an enzyme in this DH5 alpha strain that would catalyze the same reaction. Yet we have not found such activity in this strain. Moreover, the presence of mandelate at T = 0h is observed in the 3 cases presented in Fig. 5e, which led us to believe that there is a production of s-mandelate in the DH5 alpha strain of E. coli. We have indeed been able to confirm this theory, with the presence of 4-hydroxymandelate in DH1 organism, which is a similar organism to DH5 alpha [13]. What we can conclude is that there seems to be a typical (S) mandelate synthase activity , however these results have proven to be unreproducible. In order to improve our results, the use of another strain of E. coli that does not produce 4-hydroxymandelate would be a better choice. Moreover, since the results obtained are not reproducible, we could lyse bacteria before recovering the cell supernatants. Maybe there will be more products. In addition, the HPLC graph peak separation is not perfect, so a modification of the gradient parameters between solution A and B, as well as elution time, would be better.
Chitinase
Chitinase production:
We achieve to produce chitinase thanks to our construction. An inducible overproduction by pLac promoter was used to study chitinase production, induction or none-induction experiments by IPTG can show the presence of the protein. We choose to produce chitinase by E. coli K-12 DH5-alpha strain. For this two strains two differents recombinated plasmids with chit1 were tested, plasmid N°10 and N°11.
Ladder lengths for protein migration
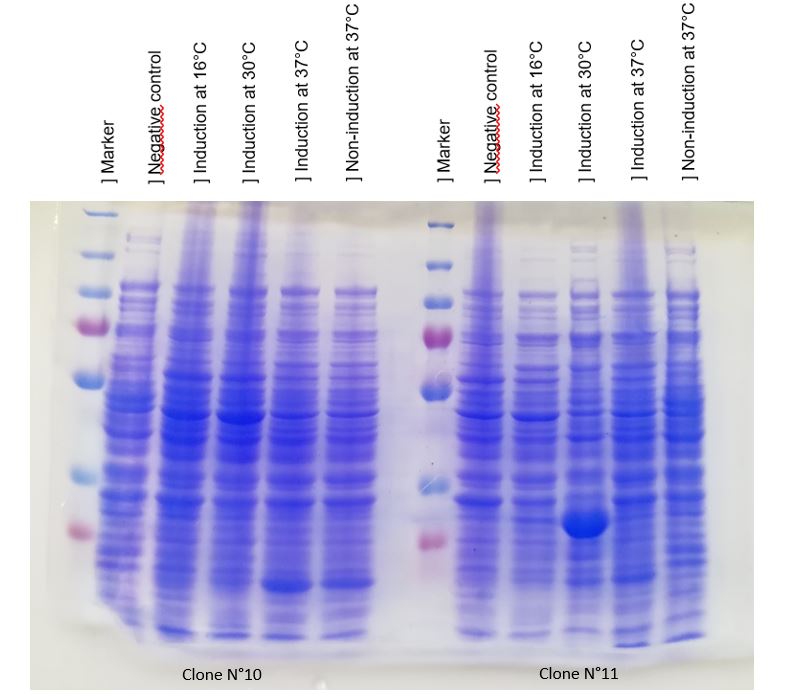
Figure 1: Figure 1: SDS-PAGE of chitinase produced by E. coli K-12 DH5-alpha.
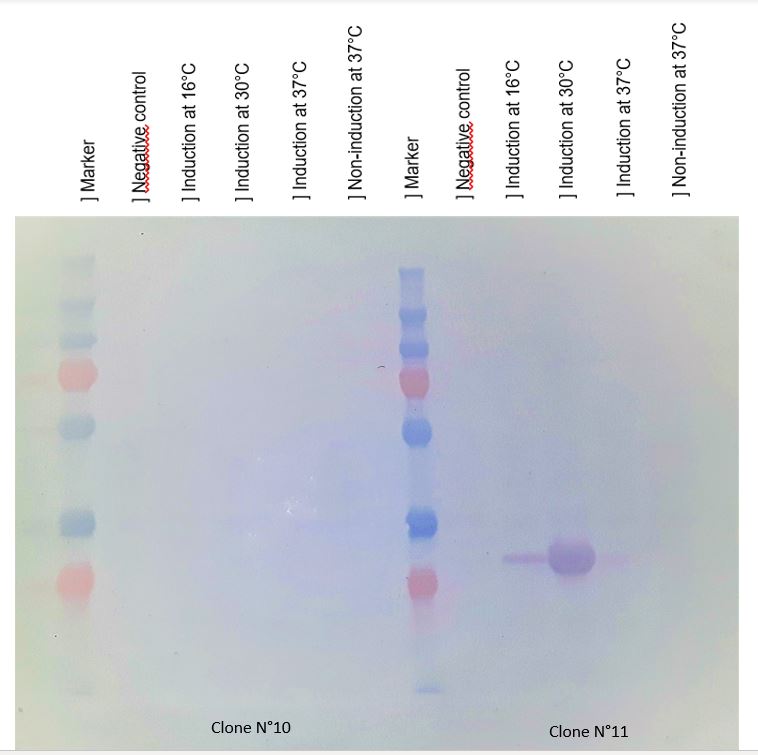
As seen in Figure 1, a protein at the expected lenght (30 kDa) was overproduced only at 30°C after IPTG induction in DH5alpha. The non-induction by IPTG show an low band of the protein overproduced. This overproduction should be the chitinase, but it isn't normal that clone N°11 wasn't able to produced at least a significant chitinase quantity at 16°C and 37°C. We clarified that thanks to a western blot, to be sure of chitinase expression by clone N°11.
Figure 2 : Western blot with anti-HIS antibodies revelation
Chitinase output are better after IPTG induction at 30°C. This condition of culture will be used to produce in large quantity chitinase for purification and activity tests.
Chitinase purification:
Chitinase designed had a polyhistidine-tag which allows it to be specifically separated among total proteins. A Akta purification was realized for purify Chit1. Akta Pure is a automated chromatography system for quick and easy purification. A nickel-nitrilotriacetic acid column was necessary to allow specifical interaction between histidines residues and metal ions from the nickel-nitrilotriacetic acid matrix.
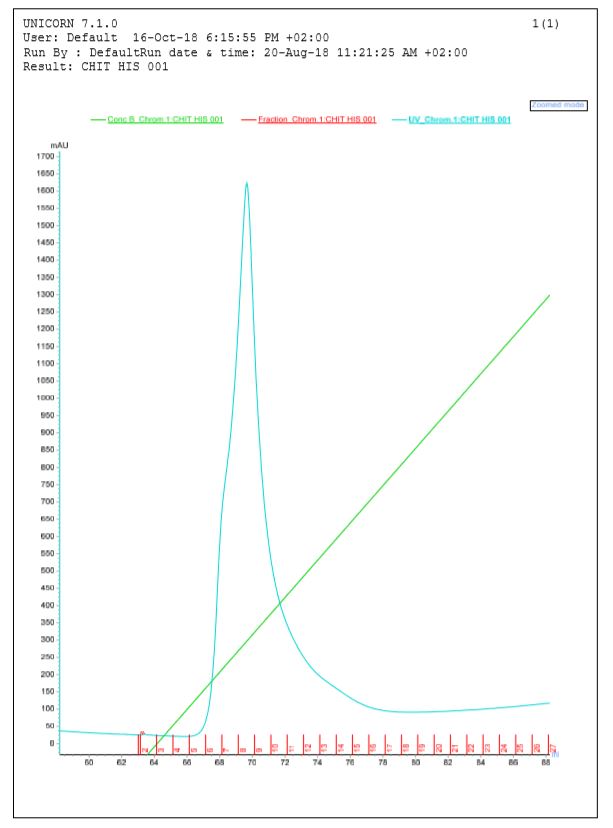
In figure 3, a elution peak corresponding to the chitinase tagged is perceptible, but a light curve at first of elution peak is commensurate with contaminants peak. There isn't a separation of these two peaks therefore contaminants proteins and protein of interest are eluated jointly in the first elution samples. According to this graph and SDS-PAGE, chitinase is present in fraction 6 to 9, but this sample weren't completely pures, a lot of nonspecific proteins were recovered. An other purification is required for better purity, in this case we didn't have time to throw again a ppurification, and that wasn't indispensible for test activity tests. But we prove here that chitinase should be purified thank to its polyhistidine tag.
Figure 3: Elution graph from chitinase purification
Figure 4: SDS-PAGE of polyhistidine-tagged chitinase purification produced by E. coli K-12 DH5-alpha, using a nickel-nitrilotriacetic acid matrix.
Showed in figure 4, a protein at 30 kDa corresponding at the chitinase is obtained on bacteria lysate, on the non-purified fraction and on fraction N°6 to fraction N°10. This fractions can be pooled and used for activity tests.
Numerous controls has been done,chitinase was produced without its peptide signal of secretion, so it will be contained in the cytoplasm, the absence of the band at 30 kDa in the pellet verify that chitinase wasn't contained in the membrane.
Bacteria lysate confirmed the presence of chitinase at 30kDa; non-purified migration shows a low quantity of chitinase with all proteins contaminants; break-though sample didn't contain the protein target demonstrating that all chitinase was fixed on nickel-nitrilotriacetic acid matrix.
The results of chitinase purification prove that chitinase can be purified thanks to its polyhistidine-tag. But purification wasn't optimal and can be ameliorated because a lot of contaminants are isolated with chitinase. The nickel-nitrilotriacetic acid matrix column used was well-worn, that can reduced its efficiency. For an optimal separation of chitinase, we should pool eluate fractions containing chitinase and purify until reduce significantly protein contaminants. But without time we continued with this purification to do activity tests.
Chitinase activity test:
Schales' procedure was used to test the chitinase activity. Schales' procedure is a colorimetric method commoly used for measuring the reducing sugars. Chitin is composed of sugar units of either N-acetyl-G-glucosamine. Chitinase degrade chitin producing N-acetyl-G-glucosamine. To monitor chitinase activity, the reducing sugars released after chitin hydrolyse are detected by a color diminution of the yellow Shales' reagent. This color diminution is translated by a absorbance diminution.
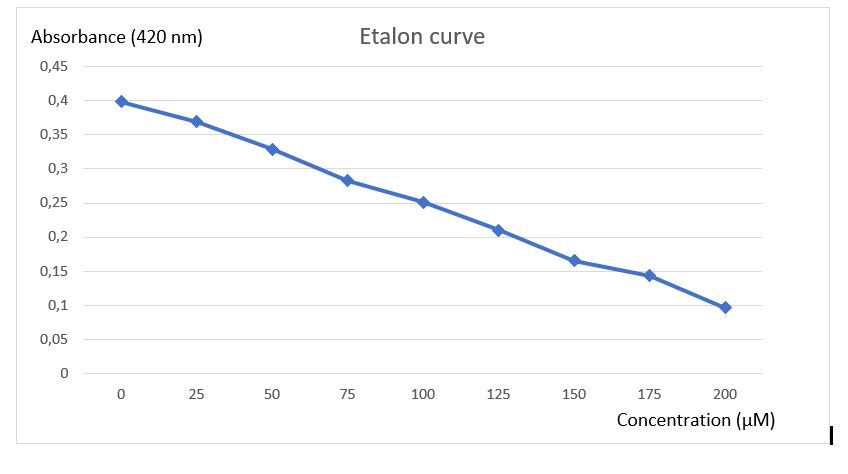
To analyse activity test results, an etalon curve have been done using the final product of chitinase reaction (N-acetyl-G-glucosamine), to know in which concentration of final product must be obtained after reaction to be detected by the method of Schales.
Figure 5: Etalon curve of N-acetyl-G-glucosamine after Schales's activity test.
A final curve has been obtain showing a absorbance diminution in function of increasing N-acetyl-G-glucosamine concentration. With this etalon curve, concentration of final product obtain after chitinase reaction can be determined. This results revealed the efficiency of Schales'procedure to detected N-acetyl-G-glucosamine to 0 at 200 uM.
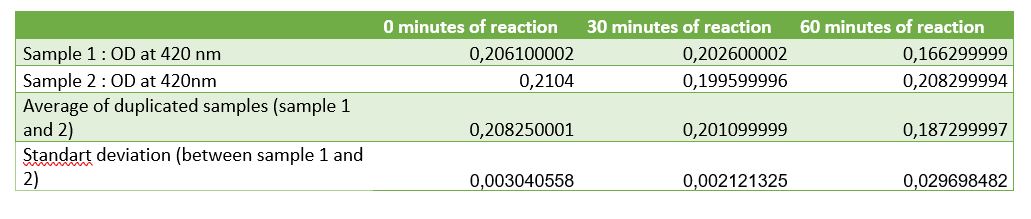
Figure 6: Table of absorbance measurements of chitinase activity test
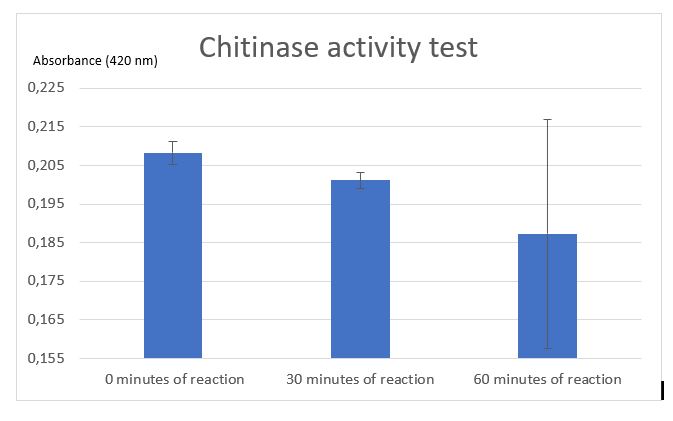
Figure 7: Chitinase activity test by Schales' procedure
According to figure 7, after 30 minutes of reaction a significant activity was mesured. But after 1 hour of reaction the standart deviation put in doubt the significant result at 30 minutes. One of the difficulties encountered was the chitin sample. Chitin sample wasn't completely dissolved in water, it was precipited and a lot of big fragments af shells of shrimps was contained making difficult the pippetage and with difficulty reproducibility. That's why an centrifugation step was added to avoid precipitated chitin to asborbe and affects the results. After centrifugation chitin wasn't really pelleted, it was easily removed. Chitin pellet should be removed in the duplicated sample of 1 hour after reaction, that can be the cause of the high standart deviation in the results. Due to limited time, we weren't able to do another activity test to improve the results and be the most careful at removing and may be we should centrigutated a longer time (not 1 minute but 5 minutes maybe).
Thanks to etalon curve if we consider only the significant results at 30 minutes, we can conclude that after 30 minutes of enzymatic reaction, our chitinase can released 125 uM of N-acetyl-G-glucosamine.
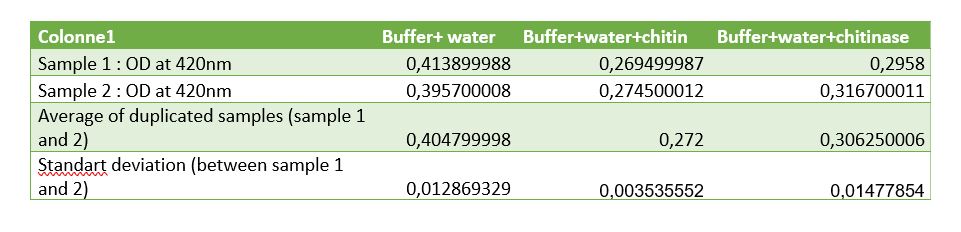
Figure 8: Table of measurements of controls for chitinase activity test
Controls were realized to be sure that the reduction of absorbance is due to chitinase reaction. A control with Schales'reagent and water show a hight absorbance by the colorimetric reactant, when chitin was added absorbance decreased that was expected because of chitin reductors components. Results of chitinase absorbance mixed with water and
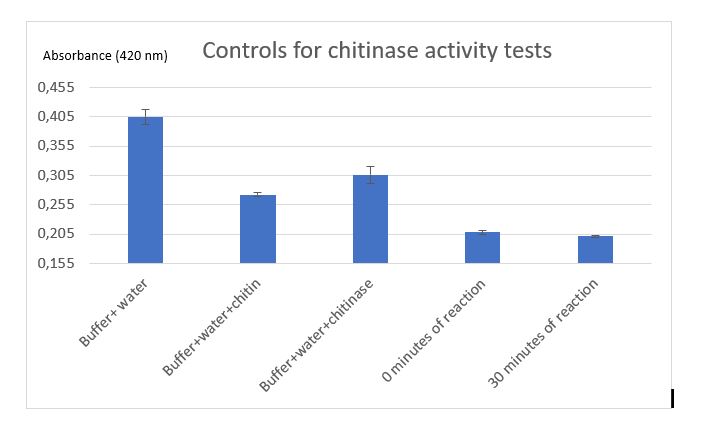
Figure 9: Controls for chitinase activity test
Trap prototype tests
(put the protocol of trap tests!!!)
Chitinase
Chitinase production:
Thanks to our construction, chitinase can be produced by E.coli strain. Chitinase production is revealed efficiency to achieve a sufficient quantity to use it to degrade chitin.
Figure 1: Figure 1: SDS-PAGE of chitinase produced by E. coli K-12 DH5-alpha.
As seen in Figure 1, a protein at the expected lenght (30 kDa) was overproduced only at 30°C after IPTG induction in DH5alpha. The non-induction by IPTG show an low band of the protein overproduced. This overproduction should be the chitinase, but it isn't normal that clone N°11 wasn't able to produced at least a significant chitinase quantity at 16°C and 37°C. We clarified that thanks to a western blot, to be sure of chitinase expression by clone N°11.
Figure 2 : Western blot with anti-HIS antibodies revelation
Chitinase output are better after IPTG induction at 30°C. This condition of culture will be used to produce in large quantity chitinase for purification and activity tests.
(put file of western and blue of BL21 or not?)
Chitinase purification:
Chitinase designed had a polyhistidine-tag which allows it to be specifically separated among total proteins. A Akta purification was realized for purify Chit1. Akta Pure is a automated chromatography system for quick and easy purification. A nickel-nitrilotriacetic acid column was necessary to allow specifical interaction between histidines residues and metal ions from the nickel-nitrilotriacetic acid matrix.
In figure 3, a elution peak corresponding to the chitinase tagged is perceptible, but a light curve at first of elution peak is commensurate with contaminants peak. There isn't a separation of these two peaks therefore contaminants proteins and protein of interest are eluated jointly in the first elution samples. According to this graph and SDS-PAGE, chitinase is present in fraction 6 to 9, but this sample weren't completely pures, a lot of nonspecific proteins were recovered. An other purification is required for better purity, in this case we didn't have time to throw again a ppurification, and that wasn't indispensible for test activity tests. But we prove here that chitinase should be purified thank to its polyhistidine tag.
Figure 3: Elution graph from chitinase purification
Figure 4: SDS-PAGE of polyhistidine-tagged chitinase purification produced by E. coli K-12 DH5-alpha, using a nickel-nitrilotriacetic acid matrix.
(put markers lenghts on the image!!!! and indicate chitinase band )
Showed in figure 4, a protein at 30 kDa corresponding at the chitinase is obtained on bacteria lysate, on the non-purified fraction and on fraction N°6 to fraction N°10. This fractions can be pooled and used for activity tests.
Numerous controls has been done,chitinase was produced without its peptide signal of secretion, so it will be contained in the cytoplasm, the absence of the band at 30 kDa in the pellet verify that chitinase wasn't contained in the membrane.
Bacteria lysate confirmed the presence of chitinase at 30kDa; non-purified migration shows a low quantity of chitinase with all proteins contaminants; break-though sample didn't contain the protein target demonstrating that all chitinase was fixed on nickel-nitrilotriacetic acid matrix.
The results of chitinase purification prove that chitinase can be purified thanks to its polyhistidine-tag. But purification wasn't optimal and can be ameliorated because a lot of contaminants are isolated with chitinase. The nickel-nitrilotriacetic acid matrix column used was well-worn, that can reduced its efficiency. For an optimal separation of chitinase, we should pool eluate fractions containing chitinase and purify until reduce significantly protein contaminants. But without time we continued with this purification to do activity tests.
Chitinase activity test:
Schales' procedure was used to test the chitinase activity. Schales' procedure is a colorimetric method commoly used for measuring the reducing sugars. Chitin is composed of sugar units of either N-acetyl-G-glucosamine. Chitinase degrade chitin producing N-acetyl-G-glucosamine. To monitor chitinase activity, the reducing sugars released after chitin hydrolyse are detected by a color diminution of the yellow Shales' reagent. This color diminution is translated by a absorbance diminution.
To analyse activity test results, an etalon curve have been done using the final product of chitinase reaction (N-acetyl-G-glucosamine), to know in which concentration of final product must be obtained after reaction to be detected by the method of Schales.
Figure 5: Etalon curve of N-acetyl-G-glucosamine after Schales's activity test.
A final curve has been obtain showing a absorbance diminution in function of increasing N-acetyl-G-glucosamine concentration. With this etalon curve, concentration of final product obtain after chitinase reaction can be determined. This results revealed the efficiency of Schales'procedure to detected N-acetyl-G-glucosamine to 0 at 200 uM.
Figure 6: Table of absorbance measurements of chitinase activity test
Figure 7: Chitinase activity test by Schales' procedure
According to figure 7, after 30 minutes of reaction a significant activity was mesured. But after 1 hour of reaction the standart deviation put in doubt the significant result at 30 minutes. One of the difficulties encountered was the chitin sample. Chitin sample wasn't completely dissolved in water, it was precipited and a lot of big fragments af shells of shrimps was contained making difficult the pippetage and with difficulty reproducibility. That's why an centrifugation step was added to avoid precipitated chitin to asborbe and affects the results. After centrifugation chitin wasn't really pelleted, it was easily removed. Chitin pellet should be removed in the duplicated sample of 1 hour after reaction, that can be the cause of the high standart deviation in the results. Due to limited time, we weren't able to do another activity test to improve the results and be the most careful at removing and may be we should centrigutated a longer time (not 1 minute but 5 minutes maybe).
Thanks to etalon curve if we consider only the significant results at 30 minutes, we can conclude that after 30 minutes of enzymatic reaction, our chitinase can released 125 uM of N-acetyl-G-glucosamine.
Figure 8: Table of measurements of controls for chitinase activity test
Controls were realized to be sure that the reduction of absorbance is due to chitinase reaction. A control with Schales'reagent, water and KPi buffer shows a hight absorbance by the colorimetric reactant, when chitin was added absorbance decreased that was expected because of chitin reductors components. Results of chitinase absorbance mixed with Schales'reagent, water and KPi buffer revealed a higher reduction absorbance at 420 nm of the enzyme but the absorbance of all the components at 0 minutes of enzymatic reaction is reliable absorbance to compare a chitinase reaction sample.
Figure 9: Controls for chitinase activity test