Kasparas12 (Talk | contribs) |
Justas2010 (Talk | contribs) |
||
| Line 36: | Line 36: | ||
<p></p> | <p></p> | ||
<p> | <p> | ||
| − | At the core of SynDrop lays a liposome. Liposomes are essentially synthetic vesicles, artificially synthesized droplets of liquid, separated from the environment by a lipid bilayer (Fig.1). They act as containers that encapsulate purified transcriptional and translational machinery and other vital elements that enable complex circuitry design. They have become increasingly popular due to various applications such as being carriers for medicinal drugs<sup>1</sup>, closed environments for protein engineering<sup>2</sup> and characterization of RNAs<sup>3</sup>, as biosensors<sup>4</sup> and molecular diagnostic tools<sup>5</sup>. The growing perspectives of liposomes as scaffolds for synthetic circuitry and membrane protein research are compelling as they have a multitude of different parameters that can be controlled. These include size, composition of a lipid membrane and interior composition. | + | At the core of SynDrop lays a liposome. Liposomes are essentially synthetic vesicles, artificially synthesized droplets of liquid, separated from the environment by a lipid bilayer (Fig. 1). They act as containers that encapsulate purified transcriptional and translational machinery and other vital elements that enable complex circuitry design. They have become increasingly popular due to various applications such as being carriers for medicinal drugs<sup>1</sup>, closed environments for protein engineering<sup>2</sup> and characterization of RNAs<sup>3</sup>, as biosensors<sup>4</sup> and molecular diagnostic tools<sup>5</sup>. The growing perspectives of liposomes as scaffolds for synthetic circuitry and membrane protein research are compelling as they have a multitude of different parameters that can be controlled. These include size, composition of a lipid membrane and interior composition. |
</p> | </p> | ||
<p> | <p> | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/c/cc/T--Vilnius-Lithuania--Fig_1_NEW_su_uzrasu_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/c/cc/T--Vilnius-Lithuania--Fig_1_NEW_su_uzrasu_Liposomes.png"/> | ||
| − | <p><strong>Fig 1</strong> The composition of a liposome with encapsulated machinery for membrane protein integration. Size, membrane composition and interior composition can be easily varied.</p> | + | <p><strong>Fig. 1</strong> The composition of a liposome with encapsulated machinery for membrane protein integration. Size, membrane composition and interior composition can be easily varied.</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 64: | Line 64: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/0/0e/T--Vilnius-Lithuania--Fig2_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/0/0e/T--Vilnius-Lithuania--Fig2_Liposomes.png"/> | ||
| − | <p><strong>Fig 2 a </strong>AutoCAD design for the photomask. There are 16 individual microchannel devices on a | + | <p><strong>Fig. 2 a </strong>AutoCAD design for the photomask. There are 16 individual microchannel devices on a |
single chip. <strong>b</strong> One device consists of three inlets, an outlet and a star-shaped junction.</p> | single chip. <strong>b</strong> One device consists of three inlets, an outlet and a star-shaped junction.</p> | ||
</p> | </p> | ||
| Line 88: | Line 88: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/7/7d/T--Vilnius-Lithuania--Fig3_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/7/7d/T--Vilnius-Lithuania--Fig3_Liposomes.png"/> | ||
| − | <p><strong>Fig 3 </strong> Simplified scheme for microfluidic device preparation. <strong>a-b</strong> the silicon wafer is cleaned and spin-coated with photoresist; <strong>c</strong> the photomask is aligned on the sample and exposed to UV light. <strong>d</strong> sample is submerged to a developer – only the sections that were exposed to the UV light remain intact on the wafer; <strong>e</strong> PDMS is poured onto the master to create a PDMS mold and left for a bake in the oven; <strong>f</strong> the mold is then separated and prepared further by cleaning and punching inlets and outlets; <strong>e-f</strong> a microscopic slide is prepared by applying a thin layer of PDMS on top; <strong>i</strong> PDMS mold and PDMS covered microscopic slide are plasma treated and connected to each other to produce a final microfluidic chip.</p> | + | <p><strong>Fig. 3 </strong> Simplified scheme for microfluidic device preparation. <strong>a-b</strong> the silicon wafer is cleaned and spin-coated with photoresist; <strong>c</strong> the photomask is aligned on the sample and exposed to UV light. <strong>d</strong> sample is submerged to a developer – only the sections that were exposed to the UV light remain intact on the wafer; <strong>e</strong> PDMS is poured onto the master to create a PDMS mold and left for a bake in the oven; <strong>f</strong> the mold is then separated and prepared further by cleaning and punching inlets and outlets; <strong>e-f</strong> a microscopic slide is prepared by applying a thin layer of PDMS on top; <strong>i</strong> PDMS mold and PDMS covered microscopic slide are plasma treated and connected to each other to produce a final microfluidic chip.</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 95: | Line 95: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/7/75/T--Vilnius-Lithuania--Fig4_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/7/75/T--Vilnius-Lithuania--Fig4_Liposomes.png"/> | ||
| − | <p><strong>Fig 4 </strong> Final form of Lipodrop.</p> | + | <p><strong>Fig. 4 </strong> Final form of Lipodrop.</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 108: | Line 108: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/2/29/T--Vilnius-Lithuania--Fig5_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/2/29/T--Vilnius-Lithuania--Fig5_Liposomes.png"/> | ||
| − | <p><strong>Fig 5 </strong> A schematic representation of the interphase of air and PVA at the star shaped junction of LipoDrop.</p> | + | <p><strong>Fig. 5 </strong> A schematic representation of the interphase of air and PVA at the star shaped junction of LipoDrop.</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 125: | Line 125: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/7/77/T--Vilnius-Lithuania--Fig6_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/7/77/T--Vilnius-Lithuania--Fig6_Liposomes.png"/> | ||
| − | <p><strong>Fig 6</strong> A close-up of the phase interface during liposome synthesis; <strong>IA</strong> phase contains elements required for the synthesis | + | <p><strong>Fig. 6</strong> A close-up of the phase interface during liposome synthesis; <strong>IA</strong> phase contains elements required for the synthesis |
and integration of membrane proteins; <strong>LO</strong> phase consists of octanol and lipids that form a lipid bilayer; OA solution | and integration of membrane proteins; <strong>LO</strong> phase consists of octanol and lipids that form a lipid bilayer; OA solution | ||
carries surfactants that stabilize the initial formation and propagation of the droplets along the microfluidic device.</p> | carries surfactants that stabilize the initial formation and propagation of the droplets along the microfluidic device.</p> | ||
| Line 142: | Line 142: | ||
</video> | </video> | ||
| − | <p><strong>Fig 7 </strong> High throughput formation of cell-sized liposomes. The video is 60x slowed down </p> | + | <p><strong>Fig. 7 </strong> High throughput formation of cell-sized liposomes. The video is 60x slowed down </p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 153: | Line 153: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/4/4e/T--Vilnius-Lithuania--Fig8_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/4/4e/T--Vilnius-Lithuania--Fig8_Liposomes.png"/> | ||
| − | <p><strong>Fig 8 </strong> An automatic detection of droplets with SpotCaliper: the droplets are marked with teal colored circles and the | + | <p><strong>Fig. 8 </strong> An automatic detection of droplets with SpotCaliper: the droplets are marked with teal colored circles and the |
diameter of each is measured; <strong>b</strong> size frequency distribution histogram fitted to Gaussian distribution (teal fit) proves | diameter of each is measured; <strong>b</strong> size frequency distribution histogram fitted to Gaussian distribution (teal fit) proves | ||
the homogeneity of the liposomes; μ=11.853 >µm±0.017 >µm ; SD=0.442 µm ±0.017 µm. <p></p> | the homogeneity of the liposomes; μ=11.853 >µm±0.017 >µm ; SD=0.442 µm ±0.017 µm. <p></p> | ||
| Line 170: | Line 170: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/c/c2/T--Vilnius-Lithuania--Fig9_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/c/c2/T--Vilnius-Lithuania--Fig9_Liposomes.png"/> | ||
| − | <p><strong>Fig 9 </strong> brightfield image of the liposomes that contain IVTT system and plasmid GFP DNA (after incubation); | + | <p><strong>Fig. 9 </strong> brightfield image of the liposomes that contain IVTT system and plasmid GFP DNA (after incubation); |
scale bar is 10 µm; <strong>b</strong> liposomes imaged with FITC: fluorescence confirms that transcription and translation | scale bar is 10 µm; <strong>b</strong> liposomes imaged with FITC: fluorescence confirms that transcription and translation | ||
reactions occur inside them; scale bar is 10 µm; <strong>c</strong> liposomes containing purified GFP protein: all the | reactions occur inside them; scale bar is 10 µm; <strong>c</strong> liposomes containing purified GFP protein: all the | ||
| Line 193: | Line 193: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/3/34/T--Vilnius-Lithuania--Fig10_Liposomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/3/34/T--Vilnius-Lithuania--Fig10_Liposomes.png"/> | ||
| − | <p><strong>Fig 10 a</strong> concentrated calcein encapsulated within liposomes: the outer solution fluoresces as some of the liposomes | + | <p><strong>Fig. 10 a</strong> concentrated calcein encapsulated within liposomes: the outer solution fluoresces as some of the liposomes |
inevitably burst releasing calcein into the outside; <strong>b</strong> box plot comparison of the control (without α-hemolysin) and | inevitably burst releasing calcein into the outside; <strong>b</strong> box plot comparison of the control (without α-hemolysin) and | ||
a group with inserted α-hemolysin; nonparametrical Mann-Whitney U test was used for the statistical evaluation: | a group with inserted α-hemolysin; nonparametrical Mann-Whitney U test was used for the statistical evaluation: | ||
| Line 321: | Line 321: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/d/d5/T--Vilnius-Lithuania--Fig4_Ribosomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/d/d5/T--Vilnius-Lithuania--Fig4_Ribosomes.png"/> | ||
| − | <p><strong> Fig. 4 </strong> PCR of homology arms, and antibiotic resistance genes</p> | + | <p><strong>Fig. 4 </strong> PCR of homology arms, and antibiotic resistance genes</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 327: | Line 327: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/8/86/T--Vilnius-Lithuania--Fig5_Ribosomes.png"/> | <img src="https://static.igem.org/mediawiki/2018/8/86/T--Vilnius-Lithuania--Fig5_Ribosomes.png"/> | ||
| − | <p><strong> Fig. 5 </strong> Constructed donor DNA sequences. The L29 donor DNA was not further revisited due to time constraints</p> | + | <p><strong>Fig. 5 </strong> Constructed donor DNA sequences. The L29 donor DNA was not further revisited due to time constraints</p> |
</p> | </p> | ||
</div> | </div> | ||
| Line 418: | Line 418: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | pRSET plasmid and Sw<sub>x</sub> PCR products were digested with restriction enzymes and ligated, while GJ<sub>x</sub> PCR products were phosphorylated and ligated to produce plasmids from linear products. DH5α competent cells were transformed and plated on lysogeny broth (LB) media with ampicillin (Amp) and grown for 16 hours. Positive colonies were selected by colony PCR or restriction analysis (Fig. 3 and Fig. 4) and grown in 5 mL LB media. Plasmids were purified and BL21 competent cells were transformed. Three tubes of every construct plus plasmid with GFP without RNA thermometer were grown till OD<sub>600</sub> reached 0.4. Control samples were taken and protein expression was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG). One tube of every construct was grown in 24 ˚C, 30 ˚C, and 37 ˚C. Samples were taken after 1 and 2 hours. SDS-PAGE was run (for elaborate protocol see <a href="https://2018.igem.org/Team:Vilnius-Lithuania/LabBook">Notebook</a>/<a href="https://2018.igem.org/Team:Vilnius-Lithuania/Protocols">Protocols</a>). Fig. 5, Fig 6 and Fig. 7 depicts GFP expression at different temperatures. Although our RNA thermometers were designed to melt at 37 ˚C, some displayed leakiness to different extent. GJ3 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622011">BBa_K2622011</a>) RNA thermometer was the leakeast and allowed for GFP translation at lower temperatures. On the other hand, when grown at 37 ˚C, it unlocked the translation of GFP to highest yields. GJ2 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622010">BBa_K2622010</a>) was less leaky, but inhibited protein translation more strictly when grown at 37 ˚C. GJ6 (link: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622012">BBa_K2622012</a>), GJ9 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622013">BBa_K2622013</a>), and GJ10 (link: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622014">BBa_K2622014</a>) suppressed GFP production at 24 ˚C and 30 ˚C at similar level. They also inhibited translation to some extent at higher temperatures, meaning their melting temperature was not reached. Altogether these results prove, that our synthetic thermoswitches are temperature-responsive and act in physiological temperature range needed for IVTT reaction and also for BamA folding and membrane insertion. | + | pRSET plasmid and Sw<sub>x</sub> PCR products were digested with restriction enzymes and ligated, while GJ<sub>x</sub> PCR products were phosphorylated and ligated to produce plasmids from linear products. DH5α competent cells were transformed and plated on lysogeny broth (LB) media with ampicillin (Amp) and grown for 16 hours. Positive colonies were selected by colony PCR or restriction analysis (Fig. 3 and Fig. 4) and grown in 5 mL LB media. Plasmids were purified and BL21 competent cells were transformed. Three tubes of every construct plus plasmid with GFP without RNA thermometer were grown till OD<sub>600</sub> reached 0.4. Control samples were taken and protein expression was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG). One tube of every construct was grown in 24 ˚C, 30 ˚C, and 37 ˚C. Samples were taken after 1 and 2 hours. SDS-PAGE was run (for elaborate protocol see <a href="https://2018.igem.org/Team:Vilnius-Lithuania/LabBook">Notebook</a>/<a href="https://2018.igem.org/Team:Vilnius-Lithuania/Protocols">Protocols</a>). Fig. 5, Fig. 6 and Fig. 7 depicts GFP expression at different temperatures. Although our RNA thermometers were designed to melt at 37 ˚C, some displayed leakiness to different extent. GJ3 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622011">BBa_K2622011</a>) RNA thermometer was the leakeast and allowed for GFP translation at lower temperatures. On the other hand, when grown at 37 ˚C, it unlocked the translation of GFP to highest yields. GJ2 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622010">BBa_K2622010</a>) was less leaky, but inhibited protein translation more strictly when grown at 37 ˚C. GJ6 (link: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622012">BBa_K2622012</a>), GJ9 (link:<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622013">BBa_K2622013</a>), and GJ10 (link: <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K2622014">BBa_K2622014</a>) suppressed GFP production at 24 ˚C and 30 ˚C at similar level. They also inhibited translation to some extent at higher temperatures, meaning their melting temperature was not reached. Altogether these results prove, that our synthetic thermoswitches are temperature-responsive and act in physiological temperature range needed for IVTT reaction and also for BamA folding and membrane insertion. |
</p> | </p> | ||
<p> | <p> | ||
| Line 577: | Line 577: | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/1/16/T--Vilnius-Lithuania--PERMATOMAS_ScFv.png" | <img src="https://static.igem.org/mediawiki/2018/1/16/T--Vilnius-Lithuania--PERMATOMAS_ScFv.png" | ||
| − | <p><strong> Fig. 1 </strong>Simplified structure of scFv Antibody</p> | + | <p><strong>Fig. 1 </strong>Simplified structure of scFv Antibody</p> |
</div> | </div> | ||
<div class="image-container"> | <div class="image-container"> | ||
<img src="https://static.igem.org/mediawiki/2018/9/97/T--Vilnius-Lithuania--Fig2_NEW_real_Surface_scFV.png" | <img src="https://static.igem.org/mediawiki/2018/9/97/T--Vilnius-Lithuania--Fig2_NEW_real_Surface_scFV.png" | ||
| − | <p><strong> Fig. 2 </strong>Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.</p> | + | <p><strong>Fig. 2 </strong>Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.</p> |
</div> | </div> | ||
<p></p> | <p></p> | ||
| Line 592: | Line 592: | ||
<div class="image-container"> | <div class="image-container"> | ||
| − | <p><strong> Fig. 3 </strong> SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.</p> | + | <p><strong>Fig. 3 </strong> SDS-PAGE of scFv. GFP is used as positive control, C- chaperone DnaK.</p> |
</div> | </div> | ||
| Line 600: | Line 600: | ||
<p> | <p> | ||
| − | <p><strong> Fig. 4 </strong> Percentage of erythrocyte lysis at different +/-scFv dilutions.</p> | + | <p><strong>Fig. 4 </strong> Percentage of erythrocyte lysis at different +/-scFv dilutions.</p> |
</div> | </div> | ||
<p>We then went one step further and constructed MstX-scFv_antiVLY <a href="http://parts.igem.org/Part:BBa_K2622038"> BBa_2622038</a>, fusion protein, aiming to increase the stability of scFv having in mind future applications and experiments of exposing it on liposome surface. Fusion protein was expressed in E.coli cells; yellow to red arrows in (Fig. 5A) indicate MstX-scFv expression after induction with IPTG.</p> | <p>We then went one step further and constructed MstX-scFv_antiVLY <a href="http://parts.igem.org/Part:BBa_K2622038"> BBa_2622038</a>, fusion protein, aiming to increase the stability of scFv having in mind future applications and experiments of exposing it on liposome surface. Fusion protein was expressed in E.coli cells; yellow to red arrows in (Fig. 5A) indicate MstX-scFv expression after induction with IPTG.</p> | ||
| Line 607: | Line 607: | ||
<p> | <p> | ||
| − | <p><strong> Fig. 5 </strong>A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.</p> | + | <p><strong>Fig. 5 </strong>A- MstX-scFv_antiVLY expression in Escherichia coli. B- scFv_antiVLY and MstX-scFv_antiVLY expression in cell-free system.</p> |
<img src="https://static.igem.org/mediawiki/2018/0/0c/T--Vilnius-Lithuania--_Fig6_Surface-scFv.png" | <img src="https://static.igem.org/mediawiki/2018/0/0c/T--Vilnius-Lithuania--_Fig6_Surface-scFv.png" | ||
| Line 613: | Line 613: | ||
<p> | <p> | ||
| − | <p><strong>Fig 6 </strong> Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.</p> | + | <p><strong>Fig. 6 </strong> Percentage of erythrocyte lysis at different scFv/MstX-scFv dilutions.</p> |
<h1>Conclusions</h1> | <h1>Conclusions</h1> | ||
<p> | <p> | ||
Revision as of 14:42, 8 November 2018
Design and Results
Results
Cell-free, synthetic biology systems open new horizons in engineering biomolecular systems which feature complex, cell-like behaviors in the absence of living entities. Having no superior genetic control, user-controllable mechanisms to regulate gene expression are necessary to successfully operate these systems. We have created a small collection of synthetic RNA thermometers that enable temperature-dependent translation of membrane proteins, work well in cells and display great potential to be transferred to any in vitro protein synthesis system.


















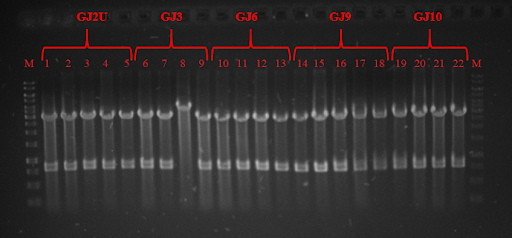









 Fig. 1 Simplified structure of scFv Antibody
Fig. 1 Simplified structure of scFv Antibody
 Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.
Fig. 2 Scheme of scFv_antiVLY and VLY interaction. Left- scFv_antiVLY binds to VLY, erythrocytes stay intact, Right- scFv_antiVLY does not bind and VLY lyse erythrocytes.



