Protocols
Contents
- 1 DH5α competent cells preparation
- 2 Cloning
- 3 Transformation
- 4 Monarch PCR and DNA cleanup kit
- 5 Monarch miniprep kit
- 6 Beauveria bassiana competent blastospores
- 7 Sabouraud medium preperation
- 8 IPTG induced protein production
- 9 Polymerase Chain Reaction (PCR)
- 10 Exponential megapriming PCR (EMP)
- 11 Protein purification
- 12 Western blot
- 13 HPLC (High Performance Liquid Chromatography) test on ...
- 14 Schales' test
- 15 Methionine-y-lyase activity test using DTNB protocol
- 16 Trap design
DH5α competent cells preparation
- The day before the preparation, make overnight starters with 100 µL of bacterial cells diluted in 5 mL of LB in a 50 mL Erlenmeyer.
- The next day, recover your starters and seed 1/60 bacterial cells in 200 mL of LB, then incubate at 37°C and 180 rpm.
- Keep measuring the OD600. When it's between 0.4 and 0.6, recover your cells and pool them in cold 50 mL Falcon tubes.
- Prepare buffers 1 and 2 and put them on ice. Put Eppendorf tubes and pipette tips at -80°C for future use.
- Centrifuge tubes at 4000 rpm and 4°C for 10 minutes.
- Discard supernatant and resuspend each Falcon tube in 20 mL of buffer 1. Afterward, centrifuge at 3500 rpm and 4°C for 5 minutes.
- Discard supernatant and resuspend each Falcon tube in 8 mL of buffer 2.
- Take 100 µL aliquots in Eppendorf tubes and stock them at -80°C.
| Product | For 40 mL |
|---|---|
| KAc (1M) | 1.2 mL |
| MnCl2(0.5M) | 4 mL |
| KCl (1M) | 4 mL |
| CaCl2 (0.1M) | 4 mL |
| Glycerol (80%) | 7.5 mL |
| H2O | 19.3 mL |
| Product | For 8.5 mL |
|---|---|
| KCl (1M) | 80 µL |
| MOPS (1M) | 400 µL |
| CaCl2 (0.1M) | 6 mL |
| Glycerol (80%) | 1.5 mL |
| H2O | 500 µL |
Cloning
Digestion
| Product | Insert | Vector |
|---|---|---|
| DNA | 1 µg | 1 µg |
| CutSmart 10X buffer | 3 µL | 3 µL |
| Restriction enzymes | 1 µL each | 1 µL each |
| MilliQ water | To 30 µL | To 30 µL |
- Incubate the mixture at 37°C for 3 hours.
- Clean-up the product using the monarch kit.
Ligation
| Product | Volume |
|---|---|
| Plasmid (Vector) | 50 ng |
| Insert | 5 x 50 ng |
| T4 DNA ligase (400 units) | 1 µL |
| T4 DNA 10X buffer | 1 µL |
| MilliQ water | To 8 µL if necessary |
- Incubate at 37°C for 5 hours.
Transformation
- Recover the needed number of competent cells tubes, adding an additional negative control tube.
- Add DNA (1µL) into the competent cells tubes.
- Incubate tubes on ice for an hour.
- Incubate tubes at 42°C for 2 minutes for thermal choc.
- Leave tubes on ice for 5 minutes.
- Leave tubes at room temperature for 5 minutes.
- Add 1 mL of LB per tube, then incubate for 1 hour at 37°C and 180 rpm.
- Spread cells on Petri dishes and incubate overnight at 37°C.
Monarch PCR and DNA cleanup kit
All centrifugation steps are done at ~13000 rpm in a typical microcentrifuge. This ensures all traces of buffer are eluted at each step.
- A starting sample volume of 50 μL is recommended. For smaller samples, nuclease-free water can be used to adjust the volume. Add 100 μL DNA Cleanup Binding Buffer to the 50 μL sample.
- Insert a column into a collection tube, load sample onto the column and close the cap. Spin for 1 minute, then discard flow-through.
- Re-insert column into the collection tube. Add 200 μL DNA Wash Buffer and spin for 1 minute. Discard flow-through.
- Repeat Step 3. This step is recommended for removal of enzymes that may interfere with downstream applications.
- Transfer column to a clean 1.5 ml microfuge tube. Use care to ensure that the tip of the column does not come into contact with the flow-through. If in doubt, re-spin for 1 minute to ensure traces of salt and ethanol are not carried over to the next step.
- Add ≥ 15 μL of hot water (70°C) to the center of the matrix. Wait for 1 minute, then spin for 1 minute to elute the DNA.
- Repeat step 6.
Monarch miniprep kit
All centrifugation steps are done at ~13000 rpm in a typical microcentrifuge. If precipitate has formed in Lysis Buffer (B2), incubate at 30–37°C, inverting periodically to dissolve. Store Plasmid Neutralization Buffer (B3) at 4°C after opening.
- Pellet 1–5 mL bacterial culture (not to exceed 15 OD units) by centrifugation for 30 seconds. Discard supernatant.
- Resuspend pellet in 200 μL Plasmid Resuspension Buffer (B1) (pink). Vortex or pipet to ensure cells are completely resuspended. There should be no visible clumps.
- Lyse cells by adding 200 μL Plasmid Lysis Buffer (B2) (blue/green). Invert the tube immediately and gently 5–6 times until color changes to dark pink and the solution is clear and viscous. Do not vortex! Incubate for 5 minutes.
- Neutralize the lysate by adding 400 μL of Plasmid Neutralization Buffer (B3) (yellow). Gently invert tube until the color is uniformly yellow and a precipitate forms. Do not vortex! Incubate for 2 minutes.
- Clarify the lysate by spinning for 10 minutes at ~13000pm.
- Carefully transfer supernatant to the spin column and centrifuge for 1 minute. Discard flow-through.
- Re-insert column in the collection tube and add 200 μL of Plasmid Wash Buffer 1. Plasmid Wash Buffer 1 removes RNA, protein, and endotoxin. Centrifuge for 1 minute. Discard flow-through.
- Add 400 μL of Plasmid Wash Buffer 2 and centrifuge for 1 minute.
- Centrifuge for 1 minute one more time.
- Transfer column to a clean 1.5 mL microfuge tube. Use care to ensure that the tip of the column has not come into contact with the flow-through. If there is any doubt, re-spin the column for 1 minute before inserting it into the clean microfuge tube.
- Add ≥ 15 μL Hot Water (70°C) to the center of the matrix. Wait for 1 minute, then spin for 1 minute to elute DNA.
- Repeat step 11.
Beauveria bassiana competent blastospores
- Dilute conidiospores in sabouraud dextrose medium and incubate for 2 days at 120 rpm and 25°C.
- Transfer 5 mL aliquots into 50 mL medium with the following products (glucose 4%, NH4NO3 0,4%, KH2PO4 0,3%, and MgSO4 0,3%) and incubate for 1 day at 120 rpm and 25°C.
- Filter conidiospores using lens paper and a steril erlenmeyer.
- Centrifuge for 5 minutes at 660 g and 4°C.
- Discard supernatant and wash in ddH2O twice by centrifugation for 5 minutes at 660 g and 4°C.
- Resuspend in 1 mL of lithium acetate (0.1M).
- Transfer into 15 mL Falcon tubes, then centrifuge for 5 minutes at 4°C and 4720 g.
- Resuspend in 0.5 mL of lithium acetate (0,1M).
- Take 100 μL aliquots in glycerol (90%).
- Stock 50 μL aliquots at -80°C.
Sabouraud medium preperation
For solid medium, mix the following products in ddH2O while maintaining the pH at 5.6:
10 g/L of mycological peptone (enzymatic digest of casein and animal tissues).
40 g/L of glucose.
15 g/L of agar.
For liquid medium, don't add agar.
IPTG induced protein production
Production
- Prepare 6 starters (20 mL LB and chloramphenicol): control BL21 (No plasmid), BL21 with the plasmid in duplicates, control DH5α (No plasmid), and DH5α with the plasmid in duplicates. Incubate overnight at 37°C and 180 rpm.
- The next day, recover the overnight starters and measure the OD600.
- Make 1/60 dilutions and incubate at 37°C to reach 0.7 in OD600.
- Add (For one of the duplicates) IPTG (1M) (10µL for 20 mL) to induce the protein production in BL21 and DH5α with the plasmid.
- Incubate at 16°C, 30°C, and 37°C for 5 hours.
- Leave the remaining cultures at 37°C for 5 hours. Incubate the cultures at 16°C overnight.
- After 5 hours, measure the OD600 and take 1 mL aliquots.
- Centrifuge aliquots for 10 minutes at 13000 g. Discard supernatant and stock samples at -20°C.
- For the remaining cultures, pour the remaining volumes into 50 mL Falcon tubes and centrifuge for 20 minutes at 4700 g and 4°C. Discard supernatant and stock samples at -20°C.
- The next day, measure the OD600 of the culture at 16°C, take 1 mL aliquots, then repeat steps 8 and 9.
Electrophoresis gel migration
- Add TSTD (1X) at 50 µL/OD unit to the 1 mL aliquots. Do not resuspend.
- Put samples in the dry bath incubator for 20 minutes.
- Prepare the migration apparel and charge the gel (20 µL for samples, and 5 µL for the ladders).
- Start migration at 0.04 A for an hour.
- Incubate gels in Coomassie blue for an hour.
- Wash gels with acetic acid till they're transparent.
- Once the gel is clear, add ddH2O and stock at 4°C.
Polymerase Chain Reaction (PCR)
Taq DNA polymerase
Setup the reaction mixture listed below on ice.
| Product | Volume |
|---|---|
| 10X reaction buffer | 2.5 µL |
| dNTPs (100 mM) | 1 µL |
| Forward primer | 1.25 µL |
| Reverse primer | 1.25 µL |
| DNA matrix | 1 µL (Whole bacteria for colony-PCR) |
| Taq DNA polymerase | 0.15 µL |
| MilliQ water | To 25 µL |
| Single cycle step (30 cycles) | Temperature | Time |
|---|---|---|
| Initial denaturation | 95°C | 10 minutes |
| Denaturation | 95°C | 30 seconds |
| Annealing | 66°C | 1 minute |
| Extension | 72°C | 1 minutes/Kb |
| Final extension | 72°C | 5 minutes |
| Hold | 16°C | Not defined |
Q5 High-Fidelity 2X master mix
| Product | For 25 µL | For 50 µL | For 100 µL |
|---|---|---|---|
| Q5 HF 2X master mix | 12.5 µL | 25 µL | 50 µL |
| Forward primer | 2.5 µL | 5 µL | 10 µL |
| Reverse primer | 2.5 µL | 5 µL | 10 µL |
| DNA matrix (10 ng/µL) | Variable | Variable | Variable |
| MilliQ water | To 25 µL | To 50 µL | To 100 µL |
| Single cycle step (30 cycles) | Temperature | Time |
|---|---|---|
| Initial denaturation | 98°C | 30 seconds |
| Denaturation | 98°C | 10 seconds |
| Annealing | Variable | 30 seconds |
| Extension | 72°C | 30 seconds/Kb |
| Final extension | 72°C | 5 minutes |
| Hold | 16°C | Not defined |
EconoTaq PLUS GREEN 2X master mix (For colony-PCR)
| Product | Volume |
|---|---|
| EconoTaq PLUS GREEN 2X master mix | 10 µL |
| Forward primer | 1 µL |
| Reverse primer | 1 µL |
| DNA matrix | Inoculate the bacteria into the PCR tube |
| Single cycle step (30 cycles) | Temperature | Time |
|---|---|---|
| Initial denaturation | 95°C | 10 minutes |
| Denaturation | 95°C | 30 seconds |
| Annealing | 60°C | 2 minutes |
| Extension | 72°C | 1 minutes/Kb |
| Final extension | 72°C | 5 minutes |
| Hold | 16°C | Not defined |
Exponential megapriming PCR (EMP)
| Product | For 100 µL | For 50 µL |
|---|---|---|
| 10X reaction buffer | 10 µL | 5 µL |
| dNTPs (100 mM) | 12 µL | 6 µL |
| DMSO | 2 µL | 1 µL |
| Forward primer | 2 µL | 1 µL |
| Reverse primer | 2 µL | 1 µL |
| DNA matrix | 1 µL | 0.5 µL |
| PfuTurbo DNA polymerase | 1 µL | 1 µL |
| MilliQ water | To 100 µL | To 50 µL |
| Single cycle step (30 cycles) | Temperature | Time |
|---|---|---|
| Initial denaturation | 95°C | 2 minutes |
| Denaturation | 95°C | 30 seconds |
| Annealing | 55°C | 1 minute |
| Extension | 44°C | 2 minutes/Kb |
| Final extension | 44°C | 20 minutes |
| Hold | 16°C | Not defined |
Digest the PCR product (To cut the methylated GATC sites) as listed below
| Product | Volume |
|---|---|
| PCR product | 50 µL |
| CutSmart 10X buffer | 6 µL |
| DpnI restriction enzyme (20 units/µL) | 2 µL |
| MilliQ water | 2 µL |
Inucubate at 37°C for 4 hours.
Protein purification
Cell lysis and protein extraction
- Prepare a 1L starter cell cultures and incubate at the optimal conditions tested out during the first protein production.
- Centrifuge for 20 minutes at 4500 rpm and 4°C . Discard supernatant.
- Resuspend pellet in a lysis buffer (50 mM Tris-HCl (pH=8), 50 mM NaCl, 1 mM EDTA, 0.1 mg/L DNase, and an anti-protease tablet).
- Put on agitator for 30 minutes at 4°C .
- Disrupt the cells using the emulsiflex.
- Ultracentrifuge for 35 minutes at 20000 rpm and 4°C .
- Transfert supernatant into 50 mL Falcon tubes and discard pellets.
His-tag Protein purification
The purification uses the ÄKTA pure system with HisTrap™ columns.
- Wash the column with 5 H2O volumes to eliminate all ethanol.
- Wash the column with 5 buffer A (50 Mm Tris-HCl (pH=8), 50 mM NaCl) volumes.
- Inject the supernatant from the protein extraction in the HisTrap™ column. Discard flowthrough.
- Elute the column with 10 buffer A volumes. Discard flowthrough.
- Using a 0 to 100% gradient, elute the column with buffer B (Buffer A + 500 mM imidazol). Collect flowthrough gradually in 1 mL fractions.
- Recover the collected fractions and verify the purification on a SDS-PAGE gel.
- Quantify the purified protein on the NanoDrop.
Protein purification by ion exchange column
The purification uses the ÄKTA pure system with anion exchange columns.
- Wash the column with 5 H2O volumes to eliminate all ethanol.
- Wash the column with 5 buffer A (50 Mm Tris-HCl (pH=8), 50 mM NaCl) volumes.
- Inject the supernatant from the protein extraction in the column. Collect flowthrough.
- Elute the column with 10 buffer A volumes. Collect flowthrough.
- Using a 0 to 100% gradient, elute the column with buffer B (50 Mm Tris-HCl (pH=8), 1 M NaCl). Collect flowthrough gradually in 1 mL fractions.
- Recover the collected fractions and verify the purification on a SDS-PAGE gel.
- Quantify the purified protein on the NanoDrop.
Western blot
Preparing a SDS-PAGE gel (4 gels recipe)
The SDS-PAGE gel has two main composites: a stacking gel and a separation gel. The following table shows the recipe to prepare them.
When putting them in the plaques pool the separation gel before the stacking gel, and leave to dry for 15 minutes.
| Product | Stacking gel | Sepration gel (12%) |
|---|---|---|
| Acrylamid | 600 µL | 6.25 mL |
| Tris-HCl (3M, pH=8.8) | 2.64 mL | - |
| Tris-HCl (1M, pH=6) | - | 600 µL |
| SDS (10%) | 50 µL | 200 µL |
| APS (10%) | 60 µL | 80 µL |
| TEMED | 5 µL | 10 µL |
When the gel is ready, prepare your samples (Dilute them in TSTD 1X at 50 µL/OD unit), then incubate for 10 minutes at 95°C.
Load the gel and run the migration system for 2 hours (10 minutes at 90V and 110 minutes at 120V).
Western blot
- Setup the migration sandwich: Whatman paper (+), nitrocellulose membrane, gel, Whatman paper (-). Soak in blotting buffer.
- Put the assembly in the blotting system and run it at 250 mA and 1 minute/kDa.
- Recover the membrane. Incubate overnight at 4°C or for an hour at RT on agitator (50 opm) in saturation buffer (TBS 1X, Tween20 0.1%, and milk 10%).
- Wash membrane in washing buffer (TBS 1X and Tween20 0.1%) for 5 minutes at 50 opm. Repeat step two more times.
- Dilute the primary antibiodies (1/1000) in TBS 1X, Tween20 0.1%, and milk 10%. Incubate membrane overnight at 4°C or an hour at RT and 20 opm.
- Wash membrane in washing buffer (TBS 1X and Tween20 0.1%) for 5 minutes at 50 opm. Repeat step two more times.
- Dilute the secondary antibiodies (1/5000) in TBS 1X, Tween20 0.1%, and milk 10%. Incubate membrane overnight at 4°C or for an hour at RT and 20 opm.
- Wash membrane in washing buffer (TBS 1X and Tween20 0.1%) for 5 minutes at 50 opm. Repeat step two more times.
HPLC (High Performance Liquid Chromatography) test on ...
Protein production
- Prepare 4 starters: control DH5α (No plasmid), DH5α with an empty pSB1C3 plasmid, DH5α with the ... pSB1C3 plasmid. Incubate overnight at 37°C.
- Make 1/60 dilutions in 50 mL (LB, glucose (20 g/L) and antibiotic when needed) and incubate at 37°C to reach 0.7 in OD600.
- Add 12,5 uL of IPTG (1M) to the cultures. Incubate overnight at 30°C.
- Prepare two stock solutions for the next day: PBS 1X, pH=7 (Solution 1) and PBS 1X (pH=7), glucose (5 g/L), phenylpyruvate (0,5 g/L) (Solution 2).
Samples preparations
- Recover overnight cultures. Centrifuge for 20 minutes at 4500 rpm and 4°C.
- Discard supernatant. Wash pellet twice with 10 mL of solution 1.
- Resuspend pellet in 50 mL of solution 2.
- Incubate at 37°C and take 1 mL aliquots every hour (for 7 hours).
Measurements
- Centrifuge 1 mL aliquots for 3 minutes at 13400 rpm.
- Take supernatant and filter it into HPLC containers.
- The HPLC system must be calibrated (use C18 column) and ready before the measurements to have fresh samples (To conserve the molecules).
Schales' test
Schales' test was used to evaluate the chitinase's functionality. The test is a colorimetric method commonly used for measuring the reducing sugars. To monitor the protein's activity, the reducing sugars released after the enzymatic reaction bound with Schales' reagent to form a yellow medium. If the protein is functional, a significant decrease in the medium's color should be observed. Equivalently, an absorbance decrease should be detected.
Buffers
- Reaction Buffer: 0,5M KPi, pH=6.
- Schales' reagent: 0,5M sodium carbonate + 0,5 g/L potassium ferricyanide.
Schales' test protocol
Three samples (at 200 µL) should be prepared, as follows, to complete the test: a blank, a sample with the substrate (Chitin), and a sample with the enzyme.
| Product | Blank | Sample 1 | Sample 2 |
|---|---|---|---|
| Reaction buffer | 20 µL | 20 µL | 20 µL |
| Substrate (Chitin) | - | 24 µL (25,3 mg/mL) | - |
| Chitinase | - | - | 5 µL |
| MilliQ water | 180 µL | 156 µL | 175 µL |
- Directly collect and add 100 µL of Schales' reagent. Then, heat sample for 15 minutes at 100°C.
- Centrifuge at top speed (~13600 rpm) in a typical microcentrifuge for 1 minute.
- Measure the absorbance (at 30°C and at 420 nm) in a plate reader (e.g TECAN).
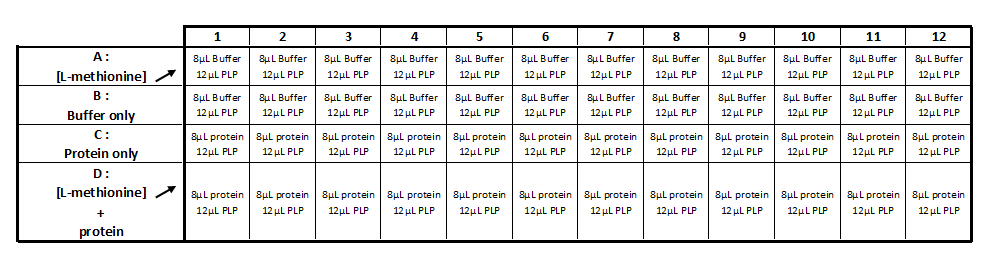
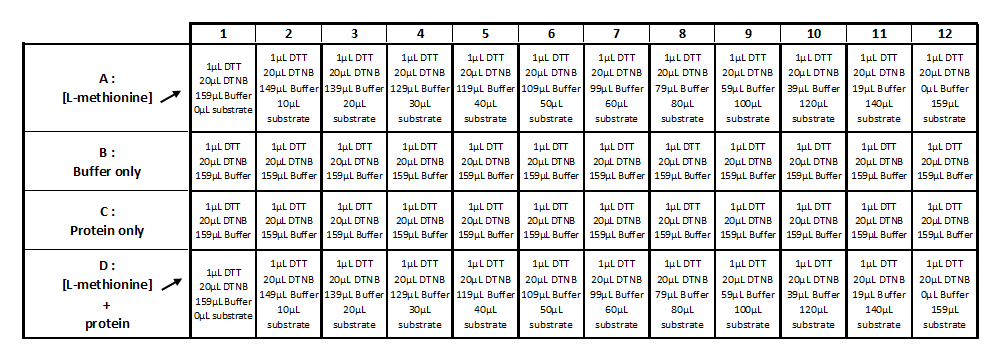
Methionine-y-lyase activity test using DTNB protocol
Solution :
- 10mM PLP
- 50mM of L-methionine in 50mM Tris-HCl pH 4
- 1mM DTT
- 1mM DTNB
- 2,5 mM Acetate de sodium pH 2,5
- 50mM Tris-HcL
- Purified protein (methionine-γ-lyase)
Realization the triplicate activity test in a 96-wells transparent plate (culture cell greiner)
Step 1 :
- Incubation during 5 min at 35 ° C: 8 μl of protein with 12 μl of the PLP solution (cf table 1)
Step 2:
- Add of 1µl of DTT
- Add of 20μL of DTNB
- Add of the substrate (cf table 2)
- QSP 200µL with 2.5mM acetate solution
- Mix and incubate during 25-30min at 25°C
Step 3 :
- Measure of OD at 320 nm with the spectrophotometer TECAN®.
Trap design
Once prototype is manufactured with cardboard.
Sample preparation protocol:
1. Beauveria bassiana sample:
-100 ug of Beauveria bassiana spores to be resuspended into 2 ml water.
2. Pheromones sample :
-DMDS and DMTS, (E)-2- hexenal, (E)-2-octenal, 2-hexanone, acetophenone, octenal, hexanone at 1.7mg/ml each of them mix in mineral oil.
Traps preparation protocol:
1) Design of sticky traps negative control without pheromones:
-Insect neutral strip and coton are sticked with 50 uL of mineral oil.
2) Design of sticky traps with pheromones:
-Insect neutral strip and coton are sticked with 50 uL of pheromone cocktail composed of DMDS and DMTS, (E)-2- hexenal, (E)-2-octenal, 2-hexanone, acetophenone, octenal, hexanone at 1.7mg/ml each of them mix in mineral oil.
3) Design of final traps testing efficiency of Beauveria bassiana:
-Insect neutral strip and coton are sticked with 50 uL of pheromone cocktail composed of pheromone cocktail at the center of the coton surrounded and separated (because DMDS and DMTS slow down fungus growth) by whole content Beauveria bassiana sample.
Test of pheromones efficiency (inspired by [1]):
-The traps are spaced 30 cm apart, and a trio of traps should be placed in three different places in the infested room.