

As a giant network of saturated hydrocarbons, polyethylene (PE) is a storehouse of a huge amount of chemical potential energy. While most microorganisms are unable to draw from this vast store as they cannot metabolise polyethylene or smaller alkanes, review of literature suggested that some microbes are capable of metabolising saturated hydrocarbons as their sole carbon source. There are however, limitations to the length of the carbon chains these microorganisms can metabolise [1].
With regards to our goal – to design a metabolism pathway for polyethylene, it was therefore a priority to fragment PE before it could be fed to the microbes. Further study on several research articles revealed that Rhodococcus ruber was among the few microorganisms that could utilize polyethylene as its main carbon source. Laccase was identified among several secreted metalloenzymes when the bacterium was grown on a PE film ([2], [6]) and so was suspected to be responsible for the cleavage effect on PE. Further analysis provided conclusive evidence that it was in fact Laccase cleaving the polyethylene backbone.
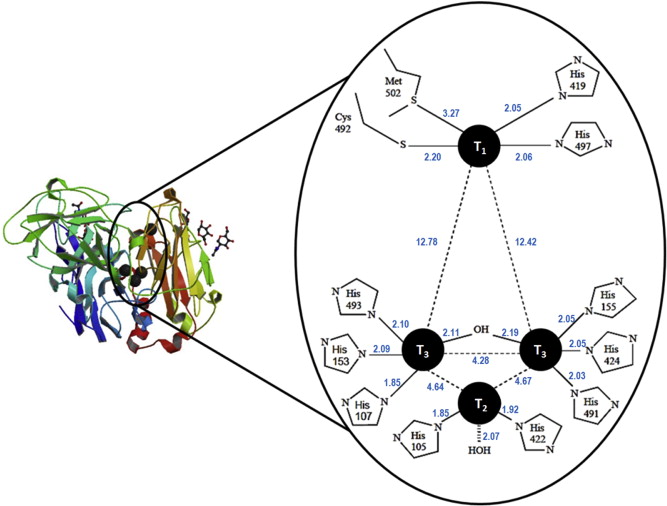
Figure 1. Protein structure and enzymatically active sites of laccase [7].
Laccase belongs to a family of enzymes known as multicopper oxidases which oxidize a variety of substrates while reducing dioxygen to water in the process. As shown in Figure 1, The enzymatically active part of laccase involves a cluster of 4 copper ions at different oxidation states [4]. The catalytic mechanism begins with the transfer of electron from substrate to the T1 copper site as a result of its higher redox potential; the electron obtained from the reduced T1 copper is then passed through the intermediate electron acceptor, T3 copper site and eventually ends up at the T2 copper site [4]. The T3 copper serves as an electron acceptor in the aerobic oxidation process while the presence of the T2 copper site is necessary as a terminal oxygen reducing site. Laccases were long reported to be present in some lignin- bio-degrading fungi, where they catalyse the oxidation, including carbonyl formation, of aromatic compounds. Nonetheless, there is considerable evidence showing laccase’s affinity to non-aromatic substrates, such as saturated hydrocarbons.
Due to the lack of any hydrolysable functional group, a specific enzymatic attack on the PE backbone may not be favourable; however, a random oxidation on PE could effectively weaken its chemical integrity. Presently, the activity of laccase on PE has been confirmed by documented cases from scientific research papers as well as from past iGEM projects. For instance, University London College IGEM team 2012 proposed that the laccase could lead to increased deterioration of polyethylene structure and was confirmed by the SEM (Scanning Electron Microscope) which showed an evident scratch made on a PE film surface after incubation with laccase, compared to the control. Moreover, in research conducted by other teams, crude laccase secreted by rhodococcus ruber was incubated with LDPE of average molecular weight 191,000. It reportedly resulted in 15 to 20 percent of mass reduction over a period of two weeks, which demonstrated the feasibility of laccase-degradation of polyethylene. To further enhance the efficiency of laccase, introduction of copper ions and mediators to the reaction medium were also justified to yield a higher degradation rate ([3], [5]) .
The target of this module was to demonstrate the ability of Laccase, expressed by E.coli, to fragment PE into simple hydrocarbons. For the purpose of utilizing laccase in the outer-membrane space, outer membrane protein A (OmpA) is required to serve as a protein signal to the membrane so that the laccase can be dumped to the intracellular space. Meanwhile, to minimise the misfolding of laccase with OmpA, our chosen OmpA includes a linker sequence at the N-termini to separate laccase from the signalling protein OmpA. To aid in protein extraction for characterization, a 6X his-tag region is added to all of our laccase. The final effective construct for activity assay is as follows:

Apart from the construct above, another construct was also involved to characterize the effectiveness of OmpA in bringing out laccase to the intracellular space.

With these two constructs constitutively expressed in E.coli, it is expected that we can compare the differences of laccase quantity in the culturing medium of E.coli for our characterisation.
REFERENCES:
1. R. Wei and W. Zimmermann, “Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we?,” Microbial Biotechnology, vol. 10, no. 6, pp. 1308–1322, 2017.
2. M. Santo, R. Weitsman, and A. Sivan, “The role of the copper-binding enzyme – laccase – in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber,” International Biodeterioration & Biodegradation, vol. 84, pp. 204–210, 2013.
3. O. V. Morozova, G. P. Shumakovich, S. V. Shleev, and Y. I. Yaropolov, “Laccase-mediator systems and their applications: A review,” Applied Biochemistry and Microbiology, vol. 43, no. 5, pp. 523–535, 2007.
4. Jones, Stephen M., and Edward I. Solomon. “Electron Transfer and Reaction Mechanism of Laccases.” Cellular and molecular life sciences : CMLS 72.5 (2015): 869–883. PMC. Web. 13 Oct. 2018.
5. Fujisawa, M., Hirai, H., & Nishida, T. Degradation of Polyethylene and Nylon-66 by the Laccase-Mediator System. Journal of Polymers and the Environment, 9(3), 103-108, 2001
6. Hadar, & Sivan. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Applied Microbiology and Biotechnology, 65(1), 97-104, 2004
7. A. Piscitelli, C. Pezzella, V. Lettera, P. Giardina, V. Faraco, and G. Sannia, “Fungal Laccases,” Fungal Enzymes, Aug. 2013.

