Demonstrate
RNA Editing as Therapy
One of the key highlights of REPAIR editing is its potential to restore pathogenic mutations back to G from A. Therefore, it is important that this technology can be used to make changes to the mRNA of disease cell lines. As such, we expanded the project to include two cancer cell lines, HeLa (a commonly used cervical cancer cell line) and MCF-7 (a commonly used breast cancer cell line) to investigate the editing efficiency of REPAIR.
The genes that we are targeting are KRAS and PPIB, which have already shown promising results in HEK293FT cells. We are particularly interested in KRAS, as KRAS is an oncogene that is closely related to tumor formation. It was found that at least one KRAS mutation is present in up to 25% of all human tumors and it is one of the most frequently activated oncogenes (Beganoyic, 2009). In another study, 33% of KRAS mutations were found to be present in combination with TP53 mutations in 480 tumors examined (Shajani-Yi et. al., 2018). As mutations in KRAS exert such a significant effect in potentially causing cancer, we feel that it is vital that we make use of REPAIR to correct the mutations of KRAS in cancer cell models.
REPAIR on Cancer Cell-Line
As such, to demonstrate the functionality of our constructs in realistic conditions, we targeted mRNA of the endogenous KRAS and PPIB genes in both HeLa and MCF-7 cell-lines to characterize the editing activities of both constructs as these are more realistic cancer cell models as compared to the HEK293FT Cells. The following figures show the editing rates of REPAIR using dCas13d-ADAR2DD and dCas13b-ADAR2DD on the two genes of PPIB and KRAS in HeLa and MCF-7 cell lines. The data for some constructs are not shown due to excessive noise in the chromatograph.


Figure 1. A-to-I editing rate of dCas13-ADAR2DD constructs targetting (a) KRAS and (b) PPIB in MCF-7 cells
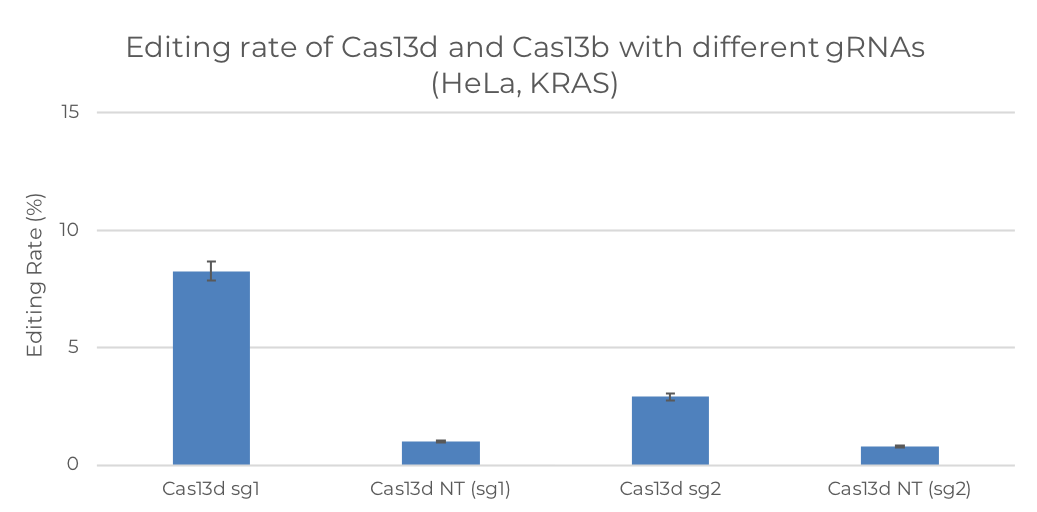

Figure 2. A-to-I editing rate of dCas13-ADAR2DD constructs targetting (a) KRAS and (b) PPIB in HeLa cells
Discussions
The editing rate for KRAS shows a general drop as compared to HEK193FT cells for all constructs. For KRAS gene in MCF-7 cell line, constructs of Cas13b show an average editing rate of above 10%, and that of Cas13d is slightly lower, of about 7%. For KRAS gene in HeLa cell line, one construct of Cas13d shows 8% editing rate. Though PPIB gene is not an important target, we can still observe that the editing rates for all constructs are consistently below 4%, except for one exceptionally high editing rate over 50% by Cas13b sg2 in HeLa cells, showing some potential for better editing activities if the guide and protein scaffold is further optimized.
Conclusion
In conclusion, though the editing rate of the cancer related oncogene KRAS plummeted from 30% - 40% in HEK293FT cells to around 10% in the two cancer cell lines, we are still able to see a clear difference between the editing rates of targeting constructs and those of non-targeting constructs, indicating that the REPAIR system and Cas13d system did edit bases from A to I in the cancer cells.
Moreover, the editing rates of the control gene, PPIB, which is not related to cancer-causing, also show a similar drop in editing efficiency. Therefore, we can conclude that the underlying cause of the drop in editing efficiency lies in the difference between HEK293FT cells and the cancer cells.
To conclude, we have demonstrated that our constructs are able to make changes to the mRNA of the cancer-causing gene KRAS in cancer cell lines HeLa and MCF-7. In order for us to make further improvement to the RNA editing ability of REPAIR, more changes have to be made to the guide RNA and the protein to enhance both its gene targeting and gene editing function.
References
1. Beganoyic, S. (2009). Clinical significance of the KRAS mutation. Bosnian journal of basic medical sciences, 9(Suppl 1), S17.
2. Shajani-Yi, Z., de Abreu, F. B., Peterson, J. D., & Tsongalis, G. J. (2018). Frequency of Somatic TP53 Mutations in Combination with Known Pathogenic Mutations in Colon Adenocarcinoma, Non–Small Cell Lung Carcinoma, and Gliomas as Identified by Next-Generation Sequencing. Neoplasia, 20(3), 256-262.
