| Line 42: | Line 42: | ||
<title>Collaborations</title> | <title>Collaborations</title> | ||
| − | + | <script> | |
| − | + | ||
| + | if(document.getElementById("home") != null) { | ||
| + | document.getElementById("homeheader").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("project") != null) { | ||
| + | document.getElementById("projecth").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("software") != null) { | ||
| + | document.getElementById("softwareh").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("parts") != null) { | ||
| + | document.getElementById("partsh").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("notebook") != null) { | ||
| + | document.getElementById("notebookh").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("human") != null) { | ||
| + | document.getElementById("humanh").classList.add('current'); | ||
| + | } | ||
| + | if(document.getElementById("people") != null) { | ||
| + | document.getElementById("peopleh").classList.add('current'); | ||
| + | } | ||
| + | </script> | ||
| + | |||
</head> | </head> | ||
| Line 49: | Line 72: | ||
| − | <body class="stretched"> | + | <body class="stretched sticky-responsive-pagemenu"> |
<!-- Document Wrapper | <!-- Document Wrapper | ||
| Line 92: | Line 115: | ||
<div class="swiper-container swiper-parent"> | <div class="swiper-container swiper-parent"> | ||
<div class="swiper-wrapper"> | <div class="swiper-wrapper"> | ||
| − | <div class="swiper-slide" style="background-image: url('images/ | + | <div class="swiper-slide" style="background-image: url('images/slider/swiper/parts2.jpg');"> |
<div class="container clearfix"> | <div class="container clearfix"> | ||
<div class="slider-caption slider-caption-center"> | <div class="slider-caption slider-caption-center"> | ||
| − | <h2 data-caption-animate="fadeInUp | + | <h2 data-caption-animate="fadeInUp">Design</h2> |
<p class="d-none d-sm-block" data-caption-animate="fadeInUp" data-caption-delay="400"></p> | <p class="d-none d-sm-block" data-caption-animate="fadeInUp" data-caption-delay="400"></p> | ||
</div> | </div> | ||
| Line 109: | Line 132: | ||
</section> | </section> | ||
| + | |||
<!-- Page Sub Menu | <!-- Page Sub Menu | ||
============================================= --> | ============================================= --> | ||
| Line 119: | Line 143: | ||
<div class="menu-title">Explore <span>DESIGN</span></div> | <div class="menu-title">Explore <span>DESIGN</span></div> | ||
| − | + | <nav class="one-page-menu"> | |
<ul> | <ul> | ||
| − | <li><a href="#" style="font-size: 90%" | + | <li><a href="#" style="font-size: 90%" |
data-href="#section-introduction">Introduction</a></li> | data-href="#section-introduction">Introduction</a></li> | ||
| − | <li><a href="#" style="font-size: 90%"data-href="#section-library | + | <li><a href="#" style="font-size: 90%" data-href="#section-library">Library</a></li> |
| − | <li><a href="#" style="font-size: 90%"data-href="#section-encapsulation">Encapsulation</a></li> | + | <li><a href="#" style="font-size: 90%" data-href="#section-encapsulation">Encapsulation</a></li> |
| − | <li><a href="#" style="font-size: 90%"data-href="#section-production">Biomolecule Production</a></li> | + | <li><a href="#" style="font-size: 90%" data-href="#section-production">Biomolecule Production</a></li> |
| − | <li><a href="#" style="font-size: 90%"data-href="#section-conversion">Substrate Conversion</a></li> | + | <li><a href="#" style="font-size: 90%" data-href="#section-conversion">Substrate Conversion</a></li> |
| − | <li><a href="#" style="font-size: 90%"data-href="#section-recording">Recording</a></li> | + | <li><a href="#" style="font-size: 90%" data-href="#section-recording">Recording</a></li> |
| − | <li><a href="#" style="font-size: 90%"data-href="#section-reading">Reading</a></li> | + | <li><a href="#" style="font-size: 90%" data-href="#section-reading">Reading</a></li> |
</ul> | </ul> | ||
</nav> | </nav> | ||
| Line 143: | Line 167: | ||
============================================= --> | ============================================= --> | ||
| − | <section | + | |
| + | <section style="overflow:hidden; position:relative; background-color: #FFF;"> | ||
| + | |||
<div class="content-wrap"> | <div class="content-wrap"> | ||
| Line 178: | Line 204: | ||
| − | <img class="image_design" src=" | + | <img class="image_design" src="https://static.igem.org/mediawiki/2018/2/27/T--Vilnius-Lithuania-OG--nucleotide_1.jpeg"> |
| Line 210: | Line 236: | ||
<p class="didesnist"><strong>General description</strong></p> | <p class="didesnist"><strong>General description</strong></p> | ||
| − | <img class="image_design_side" style="width: 60%;float: right;" src=" | + | <img class="image_design_side" style="width: 60%;float: right;" src="https://static.igem.org/mediawiki/2018/b/be/T--Vilnius-Lithuania-OG--circulization1.jpeg"> |
<p>A library of is prepared <strong>(1)</strong> for a specific or group of catalytic biomolecules - <strong>enzymes</strong>, <strong>catalytic RNAs</strong> or <strong>catalytic DNAs</strong>. For example, such library may consist of billions of randomly mutated enzyme sequences, a few computationally derived enzymes or a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="Metagenomics is the study of genetic material recovered directly from environmental samples">metagenomic</a> enzyme library.</p> | <p>A library of is prepared <strong>(1)</strong> for a specific or group of catalytic biomolecules - <strong>enzymes</strong>, <strong>catalytic RNAs</strong> or <strong>catalytic DNAs</strong>. For example, such library may consist of billions of randomly mutated enzyme sequences, a few computationally derived enzymes or a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="Metagenomics is the study of genetic material recovered directly from environmental samples">metagenomic</a> enzyme library.</p> | ||
| Line 231: | Line 257: | ||
| − | <img class="image_design" style="margin-bottom: 3%;" src=" | + | <img class="image_design" style="margin-bottom: 3%;" src="https://static.igem.org/mediawiki/2018/e/e9/T--Vilnius-Lithuania-OG--substrate.jpeg"> |
| Line 250: | Line 276: | ||
<p>Using a microfluidic device chip whole DNA library can be encapsulated into the droplets, together with specific reaction reagents. Water phase flows through the microfluidic device channels with the help of pressure created by special pumps.</p> | <p>Using a microfluidic device chip whole DNA library can be encapsulated into the droplets, together with specific reaction reagents. Water phase flows through the microfluidic device channels with the help of pressure created by special pumps.</p> | ||
| − | <img class="image_design_side" style="width: 50%; float: right" src=" | + | <img class="image_design_side" style="width: 50%; float: right" src="https://static.igem.org/mediawiki/2018/5/53/T--Vilnius-Lithuania-OG--droplet_formation.jpeg"> |
| Line 259: | Line 285: | ||
<p class="listin"><strong>3. Substrate Nucleotides</strong> - the target for catalytic biomolecule. It can be attached to any of the four <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="dCTP, dGTP, dATP, dTTP">dNTPs</a>. All four different nucleotides can be used in order to record the activity information using 4 different substrates.<br />The goal of the catalytic biomolecule is to recognise the substrate and catalyze a particular chemical conversion which depends on the substrate and biomolecules type. In many cases such conversion helps the substrate to detach from the nucleotide. Such detachment can be achieved by the first catalyzed reaction or subsequent spontaneous reactions that follow the initial one.</p> | <p class="listin"><strong>3. Substrate Nucleotides</strong> - the target for catalytic biomolecule. It can be attached to any of the four <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="dCTP, dGTP, dATP, dTTP">dNTPs</a>. All four different nucleotides can be used in order to record the activity information using 4 different substrates.<br />The goal of the catalytic biomolecule is to recognise the substrate and catalyze a particular chemical conversion which depends on the substrate and biomolecules type. In many cases such conversion helps the substrate to detach from the nucleotide. Such detachment can be achieved by the first catalyzed reaction or subsequent spontaneous reactions that follow the initial one.</p> | ||
| − | <img class="image_design" style="margin-bottom: 3%; width: 80%; margin: auto; display: block;" src=" | + | <img class="image_design" style="margin-bottom: 3%; width: 80%; margin: auto; display: block;" src="https://static.igem.org/mediawiki/2018/d/dc/T--Vilnius-Lithuania-OG--before_after.jpeg"> |
<p>Please see the Methods and Protocols section for a more detailed description of microfluidics techniques.</p> | <p>Please see the Methods and Protocols section for a more detailed description of microfluidics techniques.</p> | ||
| Line 271: | Line 297: | ||
<p>How such encapsulation looks can be seen in the video below:</p> | <p>How such encapsulation looks can be seen in the video below:</p> | ||
| + | <img style="margin-bottom: 18%; width: 100%; margin: auto; display: block;" src="https://static.igem.org/mediawiki/2018/1/1f/T--Vilnius-Lithuania-OG--encapsulation.gif"> | ||
| − | + | <p style="margin-top: 5%;">After the encapsulation each droplet on average contains single copy of a mutant DNA sequence from the Esterase library. Also every droplet contains the same amount of IVTT reagents and Substrate Nucleotides. We call these droplets the <strong>Substrate Droplets</strong>.</p> | |
| − | <p>After the encapsulation each droplet on average contains single copy of a mutant DNA sequence from the Esterase library. Also every droplet contains the same amount of IVTT reagents and Substrate Nucleotides. We call these droplets the <strong>Substrate Droplets</strong>.</p> | + | |
<div class="toggle toggle-bg"> | <div class="toggle toggle-bg"> | ||
<div class="togglet"><i class="toggle-closed icon-ok-circle"></i><i class="toggle-open icon-remove-circle"></i><strong>After encapsulation, does every droplet contain only a single DNA fragment from the library?</strong></div> | <div class="togglet"><i class="toggle-closed icon-ok-circle"></i><i class="toggle-open icon-remove-circle"></i><strong>After encapsulation, does every droplet contain only a single DNA fragment from the library?</strong></div> | ||
| Line 279: | Line 305: | ||
<p>In order to ensure the fidelity of the data collected using CAT-Seq only every 10th droplet houses a DNA template after droplet generation. If the concentration of DNA molecules in the starting encapsulation mix is too high, the likelihood of encapsulating multiple mutant templates per droplet increases drastically. Such events disrupt the data acquired by CAT-Seq or any droplet microfluidics technique. Encapsulation events can be precisely described by Poisson distribution:</p> | <p>In order to ensure the fidelity of the data collected using CAT-Seq only every 10th droplet houses a DNA template after droplet generation. If the concentration of DNA molecules in the starting encapsulation mix is too high, the likelihood of encapsulating multiple mutant templates per droplet increases drastically. Such events disrupt the data acquired by CAT-Seq or any droplet microfluidics technique. Encapsulation events can be precisely described by Poisson distribution:</p> | ||
| − | <img class="image_design" style="padding-bottom: 4%;; width: 30%; margin: auto; display: block;" src=" | + | <img class="image_design" style="padding-bottom: 4%;; width: 30%; margin: auto; display: block;" src="https://static.igem.org/mediawiki/2018/3/37/T--Vilnius-Lithuania-OG--poisson_formula.png"> |
<p>P(X = k) is the probability for single droplet to house k DNA molecules. λ shows the average number of DNA molecules per droplet. For example, if you want to have a single template per droplets, you might choose λ to be 1. The Poisson distribution predicts, that 36.7% of droplets will have one molecule. However, it also predicts that 18.4% of droplets will house 2 molecules and 6.1% - 3 molecules. Following this notation, In order to avoid artificial data, caused by multiple DNA templates in one droplet, the concentration of DNA template was kept at 1 per 10 droplets (λ = 0.1). As seen from the graph, 90.4% of droplets will be empty and 9% will have 1 template. On the other hand, 0.4%. of droplets will house 2 DNA molecules, meaning that droplets with 2 DNA templates are generated rarely</p> | <p>P(X = k) is the probability for single droplet to house k DNA molecules. λ shows the average number of DNA molecules per droplet. For example, if you want to have a single template per droplets, you might choose λ to be 1. The Poisson distribution predicts, that 36.7% of droplets will have one molecule. However, it also predicts that 18.4% of droplets will house 2 molecules and 6.1% - 3 molecules. Following this notation, In order to avoid artificial data, caused by multiple DNA templates in one droplet, the concentration of DNA template was kept at 1 per 10 droplets (λ = 0.1). As seen from the graph, 90.4% of droplets will be empty and 9% will have 1 template. On the other hand, 0.4%. of droplets will house 2 DNA molecules, meaning that droplets with 2 DNA templates are generated rarely</p> | ||
| − | <img class="image_design" style="margin-bottom: 3%; width: 100%; margin: auto; display: block;" src=" | + | <img class="image_design" style="margin-bottom: 3%; width: 100%; margin: auto; display: block;" src="https://static.igem.org/mediawiki/2018/8/89/T--Vilnius-Lithuania-OG--graph.png"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 298: | Line 324: | ||
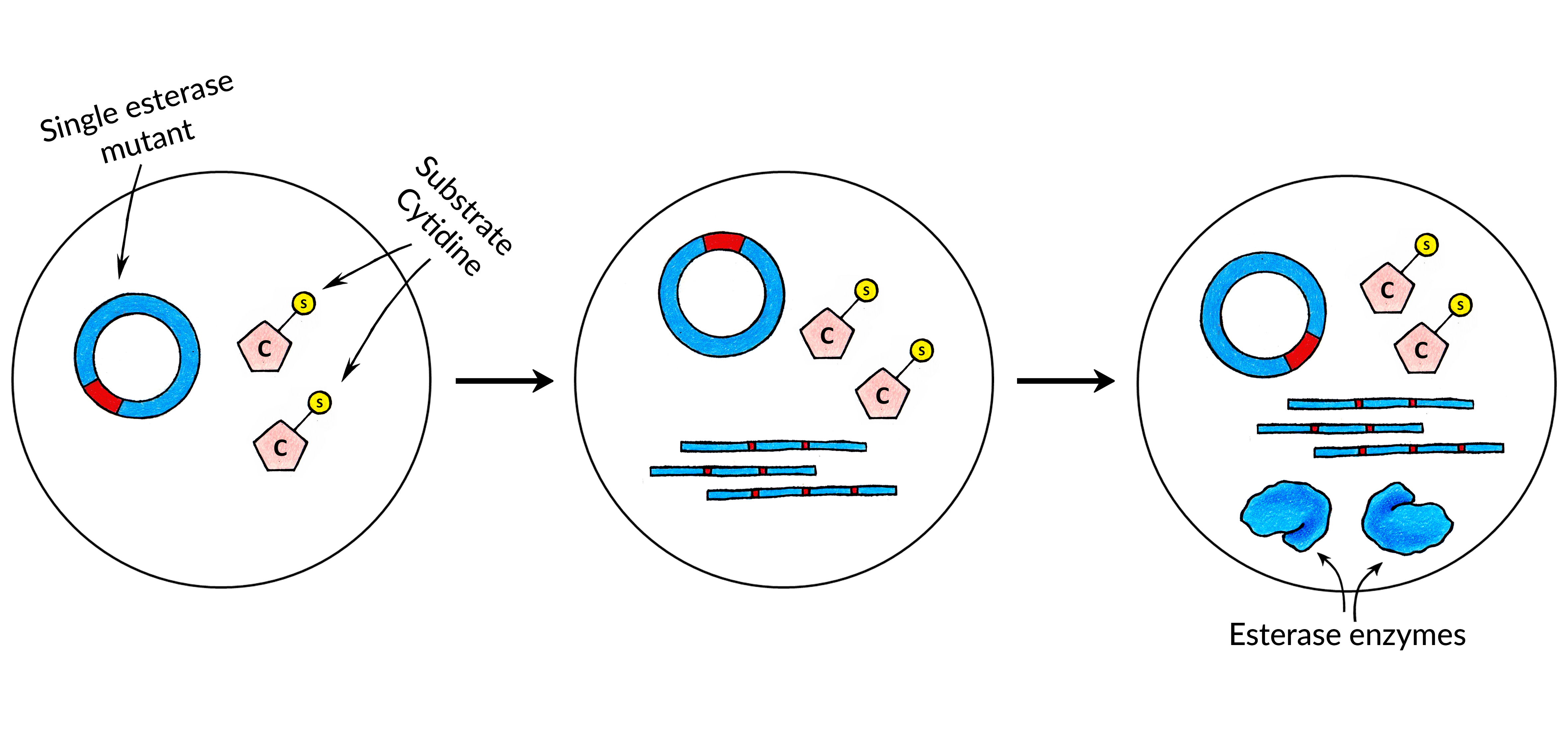
<p class="listin">1. <strong> Nucleic Acids</strong>: encapsulating DNA or RNA molecules encoding the catalytic biomolecule. After the encapsulation the droplets can be incubated in order to produce the biomolecule. If the catalytic biomolecule is an enzyme, <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="the production of protein using biological machinery in a cell-free system, that is, without the use of living cells">in-vitro protein synthesis</a> (<b>A</b>) kit can be used in order to produce it. Otherwise, if it is a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="RNA molecule capable of catalyzing a chemical reaction">Ribozyme</a> (<b>B</b>), it can be produced using <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="the production of RNA using biological machinery in a cell-free system, that is, without the use of living cells">in-vitro transcription</a> reagents. Also, there are cases where the catalytic biomolecule is a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="DNA molecule capable of catalyzing a chemical reaction">Deoxyribozyme</a> (<b>C</b>). Then, all that is required is to encapsulate the DNA library!</p> | <p class="listin">1. <strong> Nucleic Acids</strong>: encapsulating DNA or RNA molecules encoding the catalytic biomolecule. After the encapsulation the droplets can be incubated in order to produce the biomolecule. If the catalytic biomolecule is an enzyme, <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="the production of protein using biological machinery in a cell-free system, that is, without the use of living cells">in-vitro protein synthesis</a> (<b>A</b>) kit can be used in order to produce it. Otherwise, if it is a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="RNA molecule capable of catalyzing a chemical reaction">Ribozyme</a> (<b>B</b>), it can be produced using <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="the production of RNA using biological machinery in a cell-free system, that is, without the use of living cells">in-vitro transcription</a> reagents. Also, there are cases where the catalytic biomolecule is a <a href="javascript:;" data-toggle="tooltip" title="" data-original-title="DNA molecule capable of catalyzing a chemical reaction">Deoxyribozyme</a> (<b>C</b>). Then, all that is required is to encapsulate the DNA library!</p> | ||
| − | <img class="image_design" style=" margin-top: -3%; margin-bottom: 2%; display: block;" src=" | + | <img class="image_design" style=" margin-top: -3%; margin-bottom: 2%; display: block;" src="https://static.igem.org/mediawiki/2018/c/c0/T--Vilnius-Lithuania-OG--catalytic_molecules.jpeg"> |
<p> During one of the collaborative meetings with experts from microfluidic technology field in Life Sciences Center located in Vilnius, we have received information that synthesizing biomolecules from a single DNA template is an <strong>extremely inefficient process</strong>. That is because when there is only a single template of DNA, the mRNA synthesis yield is extremely low and there is <strong>not enough mRNA </strong>to make an enzyme.</p> | <p> During one of the collaborative meetings with experts from microfluidic technology field in Life Sciences Center located in Vilnius, we have received information that synthesizing biomolecules from a single DNA template is an <strong>extremely inefficient process</strong>. That is because when there is only a single template of DNA, the mRNA synthesis yield is extremely low and there is <strong>not enough mRNA </strong>to make an enzyme.</p> | ||
| − | <img class="image_design" style="margin-bottom: 3%;display: block;" src=" | + | <img class="image_design" style="margin-bottom: 3%;display: block;" src="https://static.igem.org/mediawiki/2018/9/9f/T--Vilnius-Lithuania-OG--no_protein.jpeg"> |
<p>After doing in depth literature research to try and solve this problem, we have found out that around 1990s people immensely increased short microRNA synthesis yields by circularizing DNA templates. As RNA polymerase goes around the DNA in circles it produces long strands of RNA molecules. Within those long molecules were the repeating small units of microRNA. They coined this method <strong>Rolling Circle Transcription</strong> (RCT). </p> | <p>After doing in depth literature research to try and solve this problem, we have found out that around 1990s people immensely increased short microRNA synthesis yields by circularizing DNA templates. As RNA polymerase goes around the DNA in circles it produces long strands of RNA molecules. Within those long molecules were the repeating small units of microRNA. They coined this method <strong>Rolling Circle Transcription</strong> (RCT). </p> | ||
| − | <img class="image_design" style="margin-bottom: 3%;display: block;" src=" | + | <img class="image_design" style="margin-bottom: 3%;display: block;" src="https://static.igem.org/mediawiki/2018/a/a1/T--Vilnius-Lithuania-OG--rct.jpeg"> |
<p>Following this discovery, we have figured out we can apply <strong>RCT </strong>in order to fix the current fundamental issues with biomolecule expression from single DNA templates in the field of droplet microtechnologies. As we are already circularizing DNA molecules in order to perform the MDA reaction in later steps, all that is left to do is to <strong>remove the transcription terminator</strong> from the catalytic biomolecule library members. Such removal of the terminator highly increases the mRNA concentration from a single template. Therefore, enzymes can then be <strong>efficiently synthesized</strong> using a <strong>single template of DNA</strong>.</p> | <p>Following this discovery, we have figured out we can apply <strong>RCT </strong>in order to fix the current fundamental issues with biomolecule expression from single DNA templates in the field of droplet microtechnologies. As we are already circularizing DNA molecules in order to perform the MDA reaction in later steps, all that is left to do is to <strong>remove the transcription terminator</strong> from the catalytic biomolecule library members. Such removal of the terminator highly increases the mRNA concentration from a single template. Therefore, enzymes can then be <strong>efficiently synthesized</strong> using a <strong>single template of DNA</strong>.</p> | ||
| − | <img class="image_design" style="margin-top: -2%;display: block;" src=" | + | <img class="image_design" style="margin-top: -2%;display: block;" src="https://static.igem.org/mediawiki/2018/3/3b/T--Vilnius-Lithuania-OG--rna_synthesis.jpeg"> |
| Line 319: | Line 345: | ||
| − | <img class="image_design" style="margin-top: -2%;display: block; margin-bottom: -4%" src=" | + | <img class="image_design" style="margin-top: -2%;display: block; margin-bottom: -4%" src="https://static.igem.org/mediawiki/2018/7/7f/T--Vilnius-Lithuania-OG--cells.jpeg"> |
| Line 327: | Line 353: | ||
| − | <img class="image_design" style="margin-top: -3%;display: block;" src=" | + | <img class="image_design" style="margin-top: -3%;display: block;" src="https://static.igem.org/mediawiki/2018/1/11/T--Vilnius-Lithuania-OG--long_rna.jpeg"> |
| Line 340: | Line 366: | ||
| − | <img class="image_design" style="margin-top: 0%; margin-bottom: 3%;display: block;" src=" | + | <img class="image_design" style="margin-top: 0%; margin-bottom: 3%;display: block;" src="https://static.igem.org/mediawiki/2018/3/34/T--Vilnius-Lithuania-OG--different_activity.jpeg"> |
| Line 346: | Line 372: | ||
<p>In each droplet esterase mutants are synthesized. Depending on their activities, they will remove different amount of substrates from the Substrate Cytidines in different droplets.</p> | <p>In each droplet esterase mutants are synthesized. Depending on their activities, they will remove different amount of substrates from the Substrate Cytidines in different droplets.</p> | ||
| − | <img class="image_design" style="margin-top: 0%; margin-bottom: 3%;display: block;" src=" | + | <img class="image_design" style="margin-top: 0%; margin-bottom: 3%;display: block;" src="https://static.igem.org/mediawiki/2018/c/c3/T--Vilnius-Lithuania-OG--Esterase_mutants.jpeg"> |
| Line 371: | Line 397: | ||
| − | <img class="image_design" style="margin: auto; padding-bottom: 4%; width: 70%; display: block;" src=" | + | <img class="image_design" style="margin: auto; padding-bottom: 4%; width: 70%; display: block;" src="https://static.igem.org/mediawiki/2018/8/89/T--Vilnius-Lithuania-OG--droplets_merge.jpeg"> |
| Line 379: | Line 405: | ||
| − | <img class="image_design" style="margin-top: -3%; margin-bottom: 2%; display: block;" src=" | + | <img class="image_design" style="margin-top: -3%; margin-bottom: 2%; display: block;" src="https://static.igem.org/mediawiki/2018/5/53/T--Vilnius-Lithuania-OG--dna_amplification.jpeg"> |
| Line 386: | Line 412: | ||
| − | <img class="image_design" style="margin-top: -3%; margin-bottom: 2%; display: block;" src=" | + | <img class="image_design" style="margin-top: -3%; margin-bottom: 2%; display: block;" src="https://static.igem.org/mediawiki/2018/a/ae/T--Vilnius-Lithuania-OG--amplification_2.jpeg"> |
| Line 394: | Line 420: | ||
<p style="margin-bottom: 0%;">Going back, we now have a population of <strong>substrate droplets</strong> that have a defined amount <strong>Product Nucleotides</strong> (the chemically converted substrate nucleotides). Those must then be merged with <strong>Amplification Droplets</strong>, wherein every droplet contains the same amount of amplification reagents and <strong>Reference Nucleotides</strong>. After the droplet merging, each droplet now contains a <strong>ratio of Product nucleotides to Reference Nucleotides</strong>. If the catalytic biomolecule was highly active, the ratio of Product Nucleotides to Reference Nucleotides will be high. In contrast, if the biomolecule activity was low, the ratio will also be low.</p> | <p style="margin-bottom: 0%;">Going back, we now have a population of <strong>substrate droplets</strong> that have a defined amount <strong>Product Nucleotides</strong> (the chemically converted substrate nucleotides). Those must then be merged with <strong>Amplification Droplets</strong>, wherein every droplet contains the same amount of amplification reagents and <strong>Reference Nucleotides</strong>. After the droplet merging, each droplet now contains a <strong>ratio of Product nucleotides to Reference Nucleotides</strong>. If the catalytic biomolecule was highly active, the ratio of Product Nucleotides to Reference Nucleotides will be high. In contrast, if the biomolecule activity was low, the ratio will also be low.</p> | ||
| − | <img class="image_design" style="width: 70%; display: block; margin: auto; padding-top: -5%;" src=" | + | <img class="image_design" style="width: 70%; display: block; margin: auto; padding-top: -5%;" src="https://static.igem.org/mediawiki/2018/4/42/T--Vilnius-Lithuania-OG--ratio.jpeg"> |
<p style="margin-bottom">These specific nucleotide ratios in droplets can then be <strong>recorded into DNA, by the means of DNA amplification</strong>. As DNA is amplified in each droplet, specific ratio of nucleotides is incorporated into the copy of initial template. Such amplification results in molecules that both encode the <strong>sequence of a specific biomolecule </strong>and information about that particular biomolecules’ <strong>catalytic activity</strong>.</p> | <p style="margin-bottom">These specific nucleotide ratios in droplets can then be <strong>recorded into DNA, by the means of DNA amplification</strong>. As DNA is amplified in each droplet, specific ratio of nucleotides is incorporated into the copy of initial template. Such amplification results in molecules that both encode the <strong>sequence of a specific biomolecule </strong>and information about that particular biomolecules’ <strong>catalytic activity</strong>.</p> | ||
| − | <img class="image_design" style="; display: block; margin: auto; " src=" | + | <img class="image_design" style="; display: block; margin: auto; " src="https://static.igem.org/mediawiki/2018/b/b1/T--Vilnius-Lithuania-OG--ratio_2.jpeg"> |
<p class="didesnist"><strong>Proof of concept</strong></p> | <p class="didesnist"><strong>Proof of concept</strong></p> | ||
| Line 413: | Line 439: | ||
| − | <img class="image_design" style="; display: block; margin-bottom: 2%; " src=" | + | <img class="image_design" style="; display: block; margin-bottom: 2%; " src="https://static.igem.org/mediawiki/2018/a/ac/T--Vilnius-Lithuania-OG--ratio_3.jpeg"> |
<section id="section-reading" class="page-section"> | <section id="section-reading" class="page-section"> | ||
| Line 430: | Line 456: | ||
| − | <img class="image_design" style="; display: block; margin-top: -2%; margin-bottom: 3%;" src=" | + | <img class="image_design" style="; display: block; margin-top: -2%; margin-bottom: 3%;" src="https://static.igem.org/mediawiki/2018/f/f9/T--Vilnius-Lithuania-OG--nanopore.jpeg"> |
<p style="margin-bottom: 2%">Using the same principle, billions of different catalytic biomolecule sequences and their corresponding activities can be recorded.</p> | <p style="margin-bottom: 2%">Using the same principle, billions of different catalytic biomolecule sequences and their corresponding activities can be recorded.</p> | ||
| − | <img class="image_design" style="; display: block; width: 70%; margin: auto; padding-bottom: 4%;" src=" | + | <img class="image_design" style="; display: block; width: 70%; margin: auto; padding-bottom: 4%;" src="https://static.igem.org/mediawiki/2018/c/c8/T--Vilnius-Lithuania-OG--activities_1.jpeg"> |
| Line 444: | Line 470: | ||
| − | <img class="image_design" style="; display: block; width: 80%; margin: auto; padding-bottom: 1%;" src=" | + | <img class="image_design" style="; display: block; width: 80%; margin: auto; padding-bottom: 1%;" src="https://static.igem.org/mediawiki/2018/9/93/T--Vilnius-Lithuania-OG--mutants.jpeg"> |
| Line 450: | Line 476: | ||
| − | <img class="image_design" style="; display: block; width: 80%; margin: auto; padding-bottom: 1%;" src=" | + | <img class="image_design" style="; display: block; width: 80%; margin: auto; padding-bottom: 1%;" src="https://static.igem.org/mediawiki/2018/7/73/T--Vilnius-Lithuania-OG--mut_act.jpeg"> |
| − | <p>To read about more about proof of concept results, please click here.</p> | + | <p>To read about more about proof of concept results, please <a href="https://2018.igem.org/Team:Vilnius-Lithuania-OG/Demonstrate">click here</a>.</p> |
</div> | </div> | ||
Revision as of 16:59, 17 October 2018