| (24 intermediate revisions by 4 users not shown) | |||
| Line 15: | Line 15: | ||
<h3>Design Overview</h3> | <h3>Design Overview</h3> | ||
| − | <p2>PixCell is a major improvement on an existing electrogenetic system, making it aerobic, more responsive and fully modular. The system depends on a complex network of electrochemical, chemical and biomolecular interactions.</p2> | + | <p2>PixCell is a major improvement on an existing electrogenetic system, making it aerobic, more responsive and fully modular. The system depends on a complex network of electrochemical, chemical and biomolecular interactions which is not yet fully understood. Chemical species and the cellular environment is maintained in a reduced form. Electrode potentials can either maintain this state or oxidise chemical species to activate the biological circuit. |
| + | .</p2> | ||
| − | <h3>Electrochemical Module Design</h3> | + | <h3 id="electromodule">Electrochemical Module Design</h3> |
<button class="collapsible">Overview</button> | <button class="collapsible">Overview</button> | ||
<div class="drop"> | <div class="drop"> | ||
<p2>Redox molecules are maintained in a reduced form in ambient conditions. Application of a +0.5V potential oxidises the redox molecules, allowing for activation of the genetic circuit. Application of a -0.3V potential ensures the redox molecules remain reduced, preventing activation of the genetic circuit.</p2> | <p2>Redox molecules are maintained in a reduced form in ambient conditions. Application of a +0.5V potential oxidises the redox molecules, allowing for activation of the genetic circuit. Application of a -0.3V potential ensures the redox molecules remain reduced, preventing activation of the genetic circuit.</p2> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2018/d/d6/T--Imperial_College--Electrochemicalmodulenew.png" alt="" width="50%";> | ||
</div> | </div> | ||
<button class="collapsible">Details</button> | <button class="collapsible">Details</button> | ||
| Line 29: | Line 31: | ||
<h4>Electrode or Electrode Array:</h4> | <h4>Electrode or Electrode Array:</h4> | ||
| − | <p2>The electrode is the driving force of the system. It allows for oxidation and reduction of redox molecules. </p2> | + | <p2>The electrode is the driving force of the system. It allows for oxidation and reduction of the redox molecules ferrocyanide/ferricyanide. </p2> |
<h4>Pyocyanin:</h4> | <h4>Pyocyanin:</h4> | ||
| − | <p2>This is a redox-cycling molecule produced by Pseudomonas aeuriginosa. In its oxidised form it | + | <p2>This is a redox-cycling molecule produced by <i>Pseudomonas aeuriginosa</i>. In its oxidised form it applies oxidative stress to a cell, activating redox-signalling pathways in the process. In normal aerobic conditions it is oxidised. |
| + | <div class="center"><img src="https://static.igem.org/mediawiki/2018/4/4b/T--Imperial_College--Pyostructure.png" alt="" width="20%"; ></div> | ||
</p2> | </p2> | ||
<h4>Ferrocyanide/Ferricyanide:</h4> | <h4>Ferrocyanide/Ferricyanide:</h4> | ||
| − | <p2>These molecules are well known redox mediators. | + | <p2>These molecules are well known redox mediators, meaning, they alter the redox-state of the cell. When the reduced form (ferricyanide) is present a reducing cellular environment is created, preventing the induction of redox-sensing gene circuits. When the oxidised form (ferrocyanide) is present an oxidising cellular environment is creating, permitting activation of redox-sensing gene circuit. </br><div class="center"> |
| + | <img src="https://static.igem.org/mediawiki/2018/3/38/T--Imperial_College--Ferrostructure.png" alt="" width="20%"; > | ||
| + | <img src="https://static.igem.org/mediawiki/2018/1/17/T--Imperial_College--Ferristructure.png" alt="" width="20%"; ></p2></div> | ||
<h4>Sodium Sulfite:</h4> | <h4>Sodium Sulfite:</h4> | ||
| − | <p2> | + | <p2>Sodium sulfite is known as an oxygen scavenger. This is because it reacts with oxygen and removed it from the solution. It has a minimal effect on cell growth and does not diminish fluorescence of GFP suggesting it does not fully draw oxygen out of cells when they are in a shaking incubator or on an agar plate. |
| − | + | Supposed Electrochemical Module Mechanism: | |
| − | + | Sulfite removes oxygen from solution allowing pyocyanin to be maintained in a reduced state. A potential of +0.5V generates oxidised pyocyanin and ferricyanide. Ferricyanide pushes the cell into an oxidising condition, allowing pyocyanin to remain oxidised and activate gene expression of a redox sensing gene circuit. A -0.3V potential generated reduced pyocyanin and ferrocyanide. Ferrocyanide pushes the cell into a reducing condition, allowing pyocyanin to remain reduced to prevent activation of gene expression by a redox sensing gene circuit. </br><div class="center"> | |
| − | < | + | <img src="https://static.igem.org/mediawiki/2018/4/4c/T--Imperial_College--Naso3structure.png" alt="" width="40%"; ></p2></div> |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | <h3>Biological Module Design</h3> | + | <h3 id="biomodule">Biological Module Design</h3> |
<button class="collapsible">Overview</button> | <button class="collapsible">Overview</button> | ||
<div class="drop"> | <div class="drop"> | ||
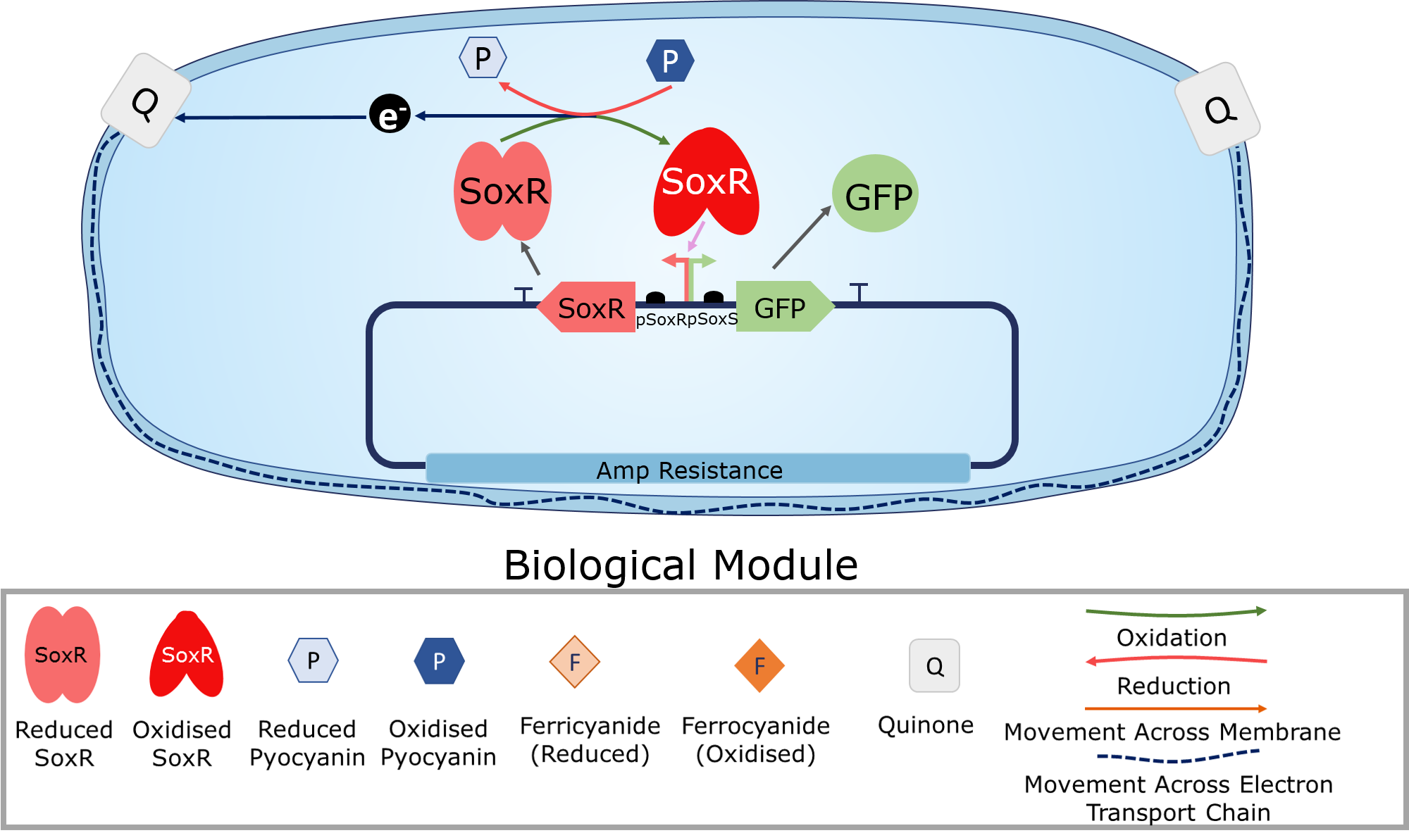
<p2>Oxidised redox molecules oxidise the transcription factor SoxR. This allows it to bind to and initiate transcription from the pSoxS promoter. This allows for the electronic induction of any gene placed downstream of this promoter.</p2> | <p2>Oxidised redox molecules oxidise the transcription factor SoxR. This allows it to bind to and initiate transcription from the pSoxS promoter. This allows for the electronic induction of any gene placed downstream of this promoter.</p2> | ||
| + | <img class="center" src="https://static.igem.org/mediawiki/2018/6/6d/T--Imperial_College--Biologicalmodulenew.png" alt="" width="50%";> | ||
</div> | </div> | ||
<button class="collapsible">Details</button> | <button class="collapsible">Details</button> | ||
<div class="drop"> | <div class="drop"> | ||
<h4>SoxR:</h4> | <h4>SoxR:</h4> | ||
| − | <p2>SoxR is a transcription factor which is constitutively expressed from the pSoxR portion of the pSoxR/pSoxS bidirectional promoter. Upon oxidation by a redox-cycling drug (such as pyocyanin) SoxR is able to activate gene transcription downstream of the pSoxS | + | <p2>SoxR is a transcription factor which is constitutively expressed from the pSoxR portion of the pSoxR/pSoxS bidirectional promoter. Upon oxidation by a redox-cycling drug (such as pyocyanin), SoxR is able to activate gene transcription downstream of the pSoxS region of the bidirectional promoter. The PixCell library contains a series of SoxR transcription factors with different activities which can be assembled modularly.</p2> |
| − | <h4> | + | <h4><i>p</i>SoxS:</h4> |
| − | <p2> | + | <p2><i>p</i>SoxS forms one half of the pSoxR/pSoxS bidirectional promoter. It is constitutively inactive, being activated by oxidised SoxR. The PixCell library includes engineered, unidirectional pSoxS promoters with variable activity which can be assembled modularly.</p2> |
<h4>Quinone Pool:</h4> | <h4>Quinone Pool:</h4> | ||
| Line 80: | Line 71: | ||
</p2> | </p2> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <h3>PixCell Electrogenetic System Design</h3> | + | <h4>Biological Module Mechanism:</h4> |
| + | When the Iron-Sulfur centres of SoxR are oxidised by oxidised pyocyanin produced by the electrochemical module, it activates transcription downstream of pSoxS. This allows for electrogenetic induction of any gene or gene circuit downstream of this promoter. | ||
| + | </div> | ||
| + | <h3 id="pixcell">PixCell Electrogenetic System Design</h3> | ||
<button class="collapsible">OFF State</button> | <button class="collapsible">OFF State</button> | ||
<div class="drop"> | <div class="drop"> | ||
| − | <p2>In ambient conditions sulfite maintains pyocyanin in a reduced state whereas ferrocyanide is stable in its reduced form. The SoxR transcription factor therefore remains reduced, preventing induction of gene transcription from the pSoxS promoter.</p2> | + | <p2>In ambient conditions or at -0.3V sulfite maintains pyocyanin in a reduced state whereas ferrocyanide is stable in its reduced form. The SoxR transcription factor therefore remains reduced, preventing induction of gene transcription from the pSoxS promoter.</p2> |
| + | <img class="center" src="https://static.igem.org/mediawiki/2018/1/19/T--Imperial_College--Offstatesystemfinal.png" alt="" width="50%";> | ||
</div> | </div> | ||
<button class="collapsible">ON State</button> | <button class="collapsible">ON State</button> | ||
<div class="drop"> | <div class="drop"> | ||
<p2>An electrode pulse of +0.5V oxidises the redox molcules pyocyanin and ferrocyanide. Pyocyanin oxidises the SoxR transcription factor which in turn initiates transcription of any gene downstream of pSoxS. This electronic induction of gene expression is amplified by the oxidised ferricyanide which pulls electrons out of the respiratory transport chain.</p2> | <p2>An electrode pulse of +0.5V oxidises the redox molcules pyocyanin and ferrocyanide. Pyocyanin oxidises the SoxR transcription factor which in turn initiates transcription of any gene downstream of pSoxS. This electronic induction of gene expression is amplified by the oxidised ferricyanide which pulls electrons out of the respiratory transport chain.</p2> | ||
| − | + | <img class="center" src="https://static.igem.org/mediawiki/2018/9/96/T--Imperial_College--Onstatesystem.png" alt="" width="50%";> | |
| + | </div> | ||
<script> | <script> | ||
Latest revision as of 18:48, 17 October 2018
Design
Design Overview
Electrochemical Module Design

Potentiostat:
Electrode or Electrode Array:
Pyocyanin:

Ferrocyanide/Ferricyanide:


Sodium Sulfite:

Biological Module Design
SoxR:
pSoxS:
Quinone Pool:
Biological Module Mechanism:
When the Iron-Sulfur centres of SoxR are oxidised by oxidised pyocyanin produced by the electrochemical module, it activates transcription downstream of pSoxS. This allows for electrogenetic induction of any gene or gene circuit downstream of this promoter.PixCell Electrogenetic System Design















