| (104 intermediate revisions by 4 users not shown) | |||
| Line 5: | Line 5: | ||
<style> | <style> | ||
.integration{ | .integration{ | ||
| − | + | margin:auto; | |
| − | + | width:70%; | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
} | } | ||
| + | |||
.integration img{ | .integration img{ | ||
| − | + | margin-top: 6%; | |
| − | + | z-index:-1; | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
} | } | ||
| − | . | + | .area{ |
| − | + | background: #A9A9A9; | |
| − | + | display:block; | |
| − | + | opacity:0; | |
| − | + | ||
| − | + | ||
} | } | ||
| − | + | ||
| − | + | #area1 :hover, #area2 :hover{ | |
| − | + | opacity:0.2; | |
| − | + | ||
| − | + | ||
} | } | ||
| + | |||
| + | .fleft{ | ||
| + | float:left; | ||
| + | clear:left; | ||
| + | width:50%; | ||
| + | text-align: center; | ||
| + | vertical-align: top; | ||
| + | display: table-cell; | ||
| + | |||
| + | } | ||
| + | .fleft p{ | ||
| + | text-align: center; | ||
| + | padding-top:6%; | ||
| + | } | ||
| + | |||
| + | .fright{ | ||
| + | float:right; | ||
| + | clear:right; | ||
| + | width:50%; | ||
| + | text-align: center; | ||
| + | display: table-cell; | ||
| + | } | ||
| + | .fright p{ | ||
| + | text-align: center; | ||
| + | padding-top:5%; | ||
| + | } | ||
| + | |||
</style> | </style> | ||
</head> | </head> | ||
| Line 51: | Line 65: | ||
</div> | </div> | ||
| − | <div class=" | + | |
| − | + | <h3>PixCell</h3> | |
| − | + | </br> | |
| + | </br> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="fleft"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/7/79/T--Imperial_College--FIGX3T.gif" alt="" width="50%"; > | ||
</div> | </div> | ||
| + | <div class="fright"> | ||
| + | <p>Electrogenetics is a synthetic biology discipline developing electronic methods to control and measure gene expression. For PixCell we developed the first aerobic electrogenetic control system.</p> | ||
| + | </div> | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <div class="row"> | |
| − | + | <div class="fleft"> | |
| − | + | ||
| − | + | <p>Using this system we demonstrated precise, programmable biological patterning using an affordable custom-built electrode array. </p> | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <div class="fright"> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/9/94/T--Imperial_College--FIGX4Tv2.gif" alt="" width="50%"; > | |
| − | + | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="row"> |
| − | + | <div class="fleft"> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/7/71/T--Imperial_College--FIGX5T.gif" alt="" width="50%"; > | |
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <div class="fright"> | ||
| + | <p>We further improved our system by building a library of electrogenetic parts compatible with a variety of assembly standards. This is the first electrogenetic toolkit and has been characterised for “plug-and-play” manipulation of the transcriptional response to electricity. </p> | ||
| − | <div class=" | + | </div> |
| − | + | ||
| − | + | </div> | |
| + | |||
| + | <div class="row"> | ||
| + | <div class="fleft"> | ||
| + | <p>Robust models of the system were developed so that electrogenetic circuits can be tested <i>in silico</i> before they are <i>in vivo</i>. | ||
| + | </p> | ||
</div> | </div> | ||
| + | <div class="fright"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/1/1c/T--Imperial_College--FIGX6T.gif" alt="" width="50%"; > | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="fleft"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/2/2e/T--Imperial_College--FIGX7Tbiocon.gif" alt="" width="90%"; > | ||
| + | </div> | ||
| + | <div class="fright"> | ||
| + | <p>Using this library we developed devices with important applications in the fields of biocontainment and manufacturing. </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="integration"> | ||
| + | |||
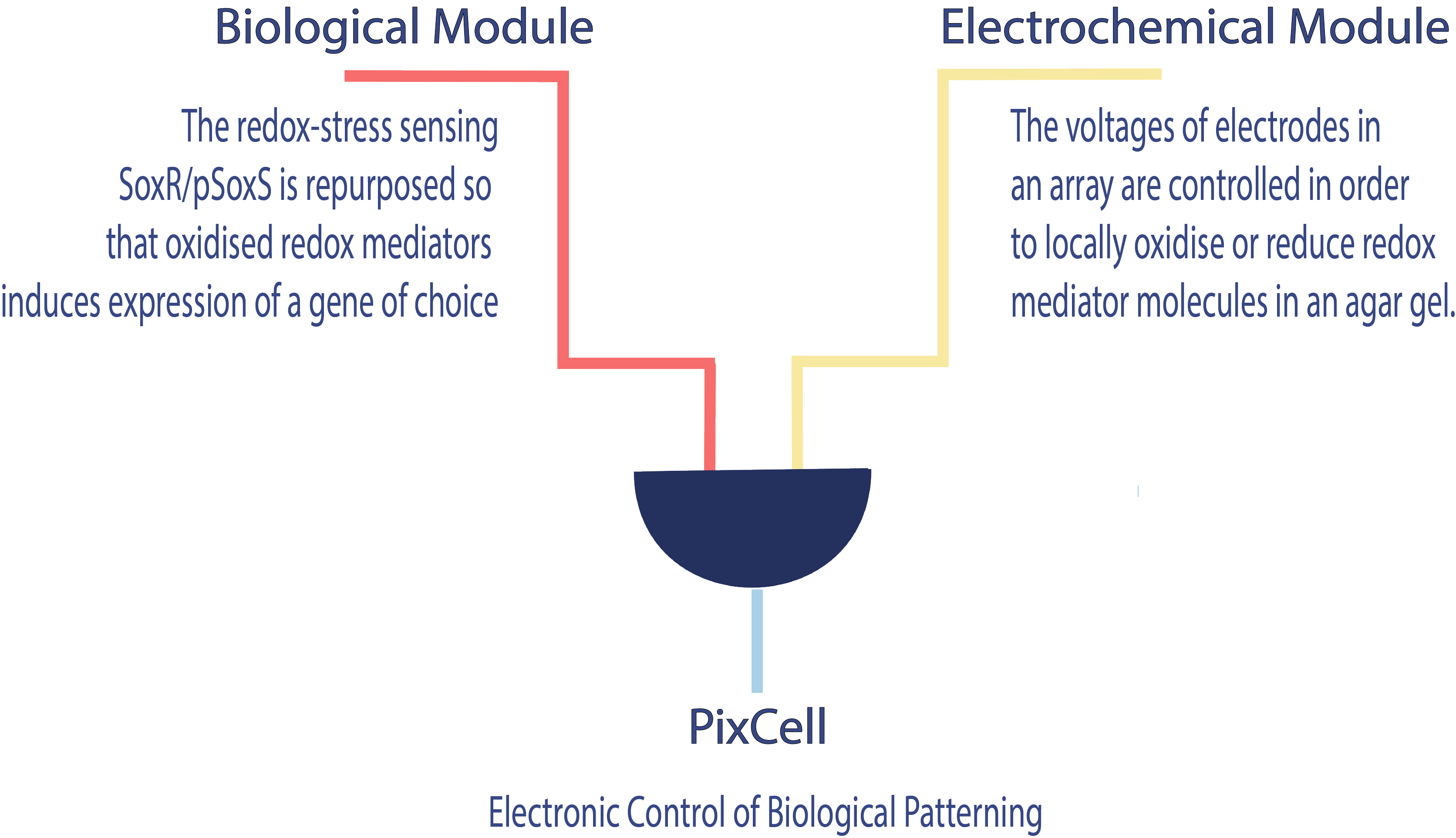
| + | <h3>Mechanism</h3> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2018/8/86/T--Imperial_College--integration.png" alt="" width="100%"; usemap="#integration" > | ||
| + | <map name="integration"> | ||
| + | <area id="area1" class="area" shape="rect" coords="100,0,350,50" href="" alt="biolmodule"> | ||
| + | <area id="area2" class="area" shape="rect" coords="550,0,1000,50" href="" alt="electromodule"> | ||
| + | <area id="area2" class="area" shape="rect" coords="" href="" alt="pixcell"> | ||
| + | </map> | ||
| + | </div> | ||
| + | |||
| + | <div class="how"> | ||
| + | <h3>Electronic control</h3> | ||
| + | <p3> The ability to control gene expression in response to electronic stimuli is a very powerful novel tool.By developing this system to work in an aerobic system, for the first time,it has brought about exciting new applications that can be further developed upon. Not only does this project introduce a new system to control gene expression to iGEM but it also expands upon the current parts available to exploit the system. Electrogenetics is, like optigenetics and the others come before it, a new arrow in the quiver of control over gene expression. | ||
| + | |||
| + | <h3>Patterning</h3> | ||
| + | <p3>Separation of labour between different cell populations allows for more complex biological processes to be engineered. Whilst Ecolibrium demonstrated a method of maintaining a stable multicellular co-culture, PixCell addresses a further necessary condition of complex multicellular life: patterning. Without patterning animals, plants and fungi would not be complex forms of life but a cellular soup. As such spatial control of gene expression is of key importance to the development of complex synthetic biology. | ||
| + | </p3> | ||
</div> | </div> | ||
| + | |||
| + | |||
</div> | </div> | ||
</body> | </body> | ||
Latest revision as of 21:49, 17 October 2018
Project Description

PixCell

Electrogenetics is a synthetic biology discipline developing electronic methods to control and measure gene expression. For PixCell we developed the first aerobic electrogenetic control system.
Using this system we demonstrated precise, programmable biological patterning using an affordable custom-built electrode array.


We further improved our system by building a library of electrogenetic parts compatible with a variety of assembly standards. This is the first electrogenetic toolkit and has been characterised for “plug-and-play” manipulation of the transcriptional response to electricity.
Robust models of the system were developed so that electrogenetic circuits can be tested in silico before they are in vivo.


Using this library we developed devices with important applications in the fields of biocontainment and manufacturing.
Mechanism
Electronic control
Patterning













