| (89 intermediate revisions by 5 users not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
<style> | <style> | ||
| + | |||
body{ | body{ | ||
background-color:#808080; | background-color:#808080; | ||
} | } | ||
| + | |||
div.naviSection { | div.naviSection { | ||
| + | } | ||
| + | img.resize { | ||
| + | max-width:50%; | ||
| + | max-height:50%; | ||
| + | display: block; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
} | } | ||
| Line 14: | Line 23: | ||
} | } | ||
| − | + | figure img { | |
| + | display: block; | ||
| + | width: 100%; | ||
| + | } | ||
| − | + | figure { | |
| − | + | padding: 5px; | |
| − | + | display: table; | |
} | } | ||
| + | |||
| + | figcaption { | ||
| + | display: table-caption; | ||
| + | caption-side: bottom; | ||
| + | padding: 0 5px 5px; | ||
| + | } | ||
| + | |||
| + | |||
| + | .w3-button:hover{color:#ff0000!important;background-color:#fff!important} | ||
| + | |||
.w3-sidebar{height:100%;width:200px;background-color:#808080;position:fixed!important;z-index:1;overflow:auto} | .w3-sidebar{height:100%;width:200px;background-color:#808080;position:fixed!important;z-index:1;overflow:auto} | ||
| Line 76: | Line 98: | ||
::-webkit-file-upload-button{-webkit-appearance:button;font:inherit} | ::-webkit-file-upload-button{-webkit-appearance:button;font:inherit} | ||
/* End extract */ | /* End extract */ | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
.w3-table,.w3-table-all{border-collapse:collapse;border-spacing:0;width:100%;display:table}.w3-table-all{border:1px solid #ccc} | .w3-table,.w3-table-all{border-collapse:collapse;border-spacing:0;width:100%;display:table}.w3-table-all{border:1px solid #ccc} | ||
.w3-bordered tr,.w3-table-all tr{border-bottom:1px solid #ddd}.w3-striped tbody tr:nth-child(even){background-color:#f1f1f1} | .w3-bordered tr,.w3-table-all tr{border-bottom:1px solid #ddd}.w3-striped tbody tr:nth-child(even){background-color:#f1f1f1} | ||
| Line 107: | Line 126: | ||
.w3-dropdown-hover.w3-mobile,.w3-dropdown-hover.w3-mobile .w3-btn,.w3-dropdown-hover.w3-mobile .w3-button,.w3-dropdown-click.w3-mobile,.w3-dropdown-click.w3-mobile .w3-btn,.w3-dropdown-click.w3-mobile .w3-button{width:100%}} | .w3-dropdown-hover.w3-mobile,.w3-dropdown-hover.w3-mobile .w3-btn,.w3-dropdown-hover.w3-mobile .w3-button,.w3-dropdown-click.w3-mobile,.w3-dropdown-click.w3-mobile .w3-btn,.w3-dropdown-click.w3-mobile .w3-button{width:100%}} | ||
.w3-button:hover{color:#000!important;background-color:#ccc!important} | .w3-button:hover{color:#000!important;background-color:#ccc!important} | ||
| − | |||
</style> | </style> | ||
| − | |||
<body> | <body> | ||
<div class = "background"> | <div class = "background"> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | < | + | <h1>Assembly Plasmids </h1> |
| − | + | ||
| − | < | + | |
<br> | <br> | ||
<!-- Sidebar --> | <!-- Sidebar --> | ||
| Line 138: | Line 150: | ||
<div class="w3-black"> | <div class="w3-black"> | ||
| − | <button id="openNav" class="w3-button w3-black w3-xlarge" onclick="w3_open()">☰</button> | + | <button id="openNav" class="w3-button w3-black w3-xlarge" onclick="w3_open()">☰<p><b>Navigate Results</b></p></button> |
| + | </div> | ||
| + | <div class="floatleft"> | ||
| + | <figure> | ||
| + | <img src= "https://static.igem.org/mediawiki/2018/9/9f/T--Austin_UTexas--KeyandPlasmid.jpeg" style="width:500px";> | ||
| + | <figcaption><b>Figure 1.</b> The nine parts that make up every Golden Gate assembly plasmid. Figure taken from "A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly" 2015 Lee, Et. al.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| − | <p> | + | <h3> Parts of an Assembly Plasmid </h3> |
| + | <p><b>Assembly plasmids contain an antibiotic resistance gene, origin of replication, origin of transfer, barcode sequence, and a 1-5 bridge part, which contains a colored reporter protein coding sequence.</b> The plasmids are made from 5 different part plasmids: a 1-5 bridge connector and part plasmids 6-8b, using a single BsaI golden gate reaction. The same promoters, terminators, and origins of transfer are used within all assemblies. Each 1-5 bridge connector has a different reporter protein coding sequence making it easy to determine which plasmid each colony contains. Each plasmid also contains a unique Barcode, that is paired to a specific origin of replication. This design eliminates as many variables as possible, allowing the origin of replication to be the single largest factor determining whether the bacteria sustain and express the plasmid. </p> | ||
| + | <div class="floatright"> | ||
| + | <figure> | ||
| + | <img src = "https://static.igem.org/mediawiki/2018/3/31/T--Austin_UTexas--RedTable.jpeg" style="width:500px";> | ||
| + | <figcaption><b>Table 1.</b> The completed assembly plasmids along with their antibiotic resistance gene, origin of replication, reporter, and barcode.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <p>We currently have 8 completed assembly plasmids and continue to make more. Table 1 shows the completed assemblies along with which reporter gene, antibiotic resistance gene, origin of replication, and barcode is on the plasmid. All assemblies have the same origin of transfer, assembly connectors, promoter/RBS, and terminator.</p> | ||
| + | <br> | ||
| + | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | |||
<br> | <br> | ||
| − | + | <br> | |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <h3> p15A Assembly Plasmid </h3> | ||
| + | <br> | ||
| + | <br> | ||
| + | <p> <b>Our lab has created a total of eight assembly plasmids and are constantly working to add more to the kit.</b> Eventually each 1-5 bridge connector, with an assigned chromoprotein reporter and barcode, will be assigned to a specific origin of replication. We currently have created three 1-5 bridge connectors with the GFP, RCP, and E2C reporters. The plate shown below demonstrates in <i> E. coli</i> cells what a successful assembly plasmid transformation looks like with a GFP reporter. While a successfully created part-plasmid does not exhibit any chromoprotein, assembly plasmids do exhibit the chromoprotein from the 1-5 bridge connector part.</p> | ||
| + | |||
| + | <div class = "floatright"> | ||
| + | <figure> | ||
| + | <img src = "https://static.igem.org/mediawiki/2018/b/b3/T--Austin_UTexas--AnnotatedCAMplate.jpg" style="width:300px";> | ||
| + | <figcaption><b>Figure 2. Assembly plasmid BHR904</b> This plasmid contains the following parts: 901.1 Bridge connector (GFP), CAM 8b antibiotic resistance, p15A origin of replication, barcode 3, and the origin of transfer. | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | </div> | ||
| + | |||
| + | <p> In figure 2, assembly BHR904 has the 1-5 bridge connector with GFP (901.1) as you can see the single green fluorescent colony on a plate of other non-fluorescent colonies.These non-fluorescent colonies did not pick up the BHR904 plasmid but still have CAM resistance possibly from picking up other undigested part plasmids (all part plasmids have CAM resistance). It is normal to only see a few colony that are transformed with the assembly plasmid and express the chromoprotein</p> | ||
| + | |||
| + | <h3> Shuttle Vector Assembly</h3> | ||
| + | |||
| + | <div class = "floatleft"> | ||
| + | <figure> | ||
| + | <img src = "https://static.igem.org/mediawiki/2018/e/e2/T--Austin_UTexas--ShuttleVectorAssembly5.jpg" style="width:300px";> | ||
| + | <figcaption><b>Figure 3.</b> BHR 925 Golden Gate Assembly</b> | ||
<br> | <br> | ||
| − | + | BHR 925 assembly on a LB + CRB plate, viewed under blue light. Red fluorescent colonies (circled) were barely visible after a day of growth but more pronounced after two days. | |
| − | + | ||
| − | + | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | </div> | ||
| + | |||
| + | <p> Broad Host Range assemblies encoding a shuttle vector, pAMBI, were assembled using standard Golden Gate assembly protocols. BHR 925 (E2-Crimson + pAMBI Origin + CRB resistance), 920 (E2-Crimson + pAMBI Origin + KAN resistance), and 922 produced darker, bluish colonies under visible light. When viewed under a UV light, the colonies were red, indicating the successful expression of E2-Crimson | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | |||
<p>Minpreps of the BHR 922 assemby, which produced colonies but was shown to be only the E2-Crimson | <p>Minpreps of the BHR 922 assemby, which produced colonies but was shown to be only the E2-Crimson | ||
1 – 5 bridge, was re-transformed into E. coli along with BHR 920 and BHR 925 to better view expression</p> | 1 – 5 bridge, was re-transformed into E. coli along with BHR 920 and BHR 925 to better view expression</p> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | + | <div class = "floatright"> | |
| − | + | <figure> | |
| − | + | <img src = "https://static.igem.org/mediawiki/2018/4/43/T--Austin_UTexas--shuttlevectorfigure.png" style="width:700px";> | |
| + | <figcaption><b>Figure 4.</b> BHR 925 Shuttle Vector Assembly Transformed into Top 10. Viewed under visible light, the dark blue colonies are clearly present (left). Shuttle Vector Assembly Minipreps Transformed into Top 10 (right). | ||
| + | <br> | ||
| + | Plate 1: E2-Crimson 1-5 Bridge plated on a LB + CAM plate<br> | ||
| + | Plate 2: BHR 925 assembly on a LB + CRB plate<br> | ||
| + | Plate 3: BHR 922 assembly plated on a LB + KAN plate | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| + | <script> | ||
| + | function w3_open() { | ||
| + | document.getElementById("main").style.marginLeft = "25%"; | ||
| + | document.getElementById("mySidebar").style.width = "25%"; | ||
| + | document.getElementById("mySidebar").style.display = "block"; | ||
| + | document.getElementById("openNav").style.display = 'none'; | ||
| + | } | ||
| + | function w3_close() { | ||
| + | document.getElementById("main").style.marginLeft = "0%"; | ||
| + | document.getElementById("mySidebar").style.display = "none"; | ||
| + | document.getElementById("openNav").style.display = "inline-block"; | ||
| + | } | ||
| + | </script> | ||
| + | </body> | ||
</html> | </html> | ||
Latest revision as of 03:45, 18 October 2018
Assembly Plasmids

Parts of an Assembly Plasmid
Assembly plasmids contain an antibiotic resistance gene, origin of replication, origin of transfer, barcode sequence, and a 1-5 bridge part, which contains a colored reporter protein coding sequence. The plasmids are made from 5 different part plasmids: a 1-5 bridge connector and part plasmids 6-8b, using a single BsaI golden gate reaction. The same promoters, terminators, and origins of transfer are used within all assemblies. Each 1-5 bridge connector has a different reporter protein coding sequence making it easy to determine which plasmid each colony contains. Each plasmid also contains a unique Barcode, that is paired to a specific origin of replication. This design eliminates as many variables as possible, allowing the origin of replication to be the single largest factor determining whether the bacteria sustain and express the plasmid.
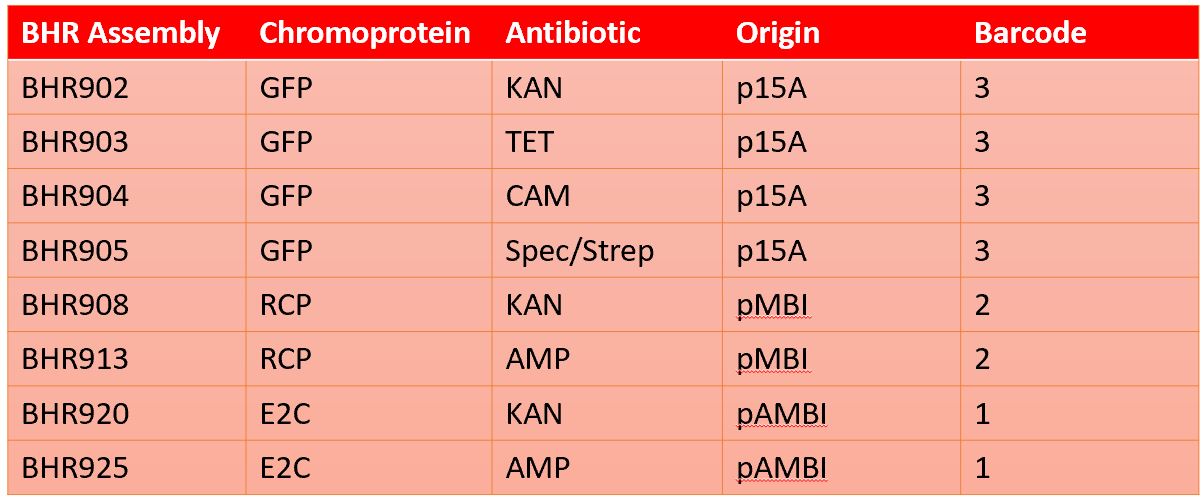
We currently have 8 completed assembly plasmids and continue to make more. Table 1 shows the completed assemblies along with which reporter gene, antibiotic resistance gene, origin of replication, and barcode is on the plasmid. All assemblies have the same origin of transfer, assembly connectors, promoter/RBS, and terminator.
p15A Assembly Plasmid
Our lab has created a total of eight assembly plasmids and are constantly working to add more to the kit. Eventually each 1-5 bridge connector, with an assigned chromoprotein reporter and barcode, will be assigned to a specific origin of replication. We currently have created three 1-5 bridge connectors with the GFP, RCP, and E2C reporters. The plate shown below demonstrates in E. coli cells what a successful assembly plasmid transformation looks like with a GFP reporter. While a successfully created part-plasmid does not exhibit any chromoprotein, assembly plasmids do exhibit the chromoprotein from the 1-5 bridge connector part.

In figure 2, assembly BHR904 has the 1-5 bridge connector with GFP (901.1) as you can see the single green fluorescent colony on a plate of other non-fluorescent colonies.These non-fluorescent colonies did not pick up the BHR904 plasmid but still have CAM resistance possibly from picking up other undigested part plasmids (all part plasmids have CAM resistance). It is normal to only see a few colony that are transformed with the assembly plasmid and express the chromoprotein
Shuttle Vector Assembly

BHR 925 assembly on a LB + CRB plate, viewed under blue light. Red fluorescent colonies (circled) were barely visible after a day of growth but more pronounced after two days.
Broad Host Range assemblies encoding a shuttle vector, pAMBI, were assembled using standard Golden Gate assembly protocols. BHR 925 (E2-Crimson + pAMBI Origin + CRB resistance), 920 (E2-Crimson + pAMBI Origin + KAN resistance), and 922 produced darker, bluish colonies under visible light. When viewed under a UV light, the colonies were red, indicating the successful expression of E2-Crimson
Minpreps of the BHR 922 assemby, which produced colonies but was shown to be only the E2-Crimson 1 – 5 bridge, was re-transformed into E. coli along with BHR 920 and BHR 925 to better view expression

Plate 1: E2-Crimson 1-5 Bridge plated on a LB + CAM plate
Plate 2: BHR 925 assembly on a LB + CRB plate
Plate 3: BHR 922 assembly plated on a LB + KAN plate
