AzirQuantum (Talk | contribs) |
XZR20020416 (Talk | contribs) |
||
| (21 intermediate revisions by 4 users not shown) | |||
| Line 6: | Line 6: | ||
<body> | <body> | ||
<link id="Sec_CSS" rel="stylesheet" type="text/css" href="https://2018.igem.org/Template:SHSBNU_China/Sample_css?action=raw&ctype=text/css" /> | <link id="Sec_CSS" rel="stylesheet" type="text/css" href="https://2018.igem.org/Template:SHSBNU_China/Sample_css?action=raw&ctype=text/css" /> | ||
| − | <script src=" | + | <script src="http://22018.igem.org/Template:SHSBNU_China/Sample_js?action=raw&ctype=text/javascript" type="text/javascript"></script> |
<div id="main_main"> | <div id="main_main"> | ||
<section id="main"> | <section id="main"> | ||
| Line 93: | Line 93: | ||
<div id="menu"> | <div id="menu"> | ||
| − | < | + | <h6 id="menu_intro">Experiment</h6> |
<div class="second_classfication"> | <div class="second_classfication"> | ||
<a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/Experiments#Outline">Outline</a> | <a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/Experiments#Outline">Outline</a> | ||
| Line 108: | Line 108: | ||
<div class="second_classfication"> | <div class="second_classfication"> | ||
| − | <a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/ | + | <a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/Protocol">Protocol</a> |
</div> | </div> | ||
<div class="second_classfication"> | <div class="second_classfication"> | ||
<a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/InterLab">InterLab</a> | <a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/InterLab">InterLab</a> | ||
| + | </div> | ||
| + | |||
| + | <div class="second_classfication"> | ||
| + | <a class="snd_class" href="https://2018.igem.org/Team:SHSBNU_China/Applied_Design">Applied Design</a> | ||
</div> | </div> | ||
| Line 131: | Line 135: | ||
<p class="text"> | <p class="text"> | ||
| − | Our system includes two main | + | Our system includes two main parts one is CotA laccase and the other is biofilm containing SpyTag. So our experiments were mainly divided in three periods, verifying both parts individually and verifying Biofilm x Laccase system as a whole. |
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | The first period was to construct our biofilm | + | The first period was to construct our biofilm containing SpyTag. We fused SpyTag to the CsgA, which is the major component of our biofilm. We successfully verified that protein fused with spycatcher domain could be fixed on our biofilm. |
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | + | Then we isolated CotA gene of B. subtilis WT168 and successfully expressed this laccase in E. coli cell. We confirmed its oxidation function. Furthermore, we attached CotA coding sequence with several signal peptides and tested the efficiency of secretion. </p> | |
| − | + | ||
<p class="text"> | <p class="text"> | ||
| − | The final period is mainly focused on combination of | + | The final period is mainly focused on combination of the two parts above. Experiments were conducted to measure the laccase activity of the total Biofilm x Laccase system. A significant difference was confirmed between the experiment group and the control group. Thus, our system is successful. |
</p> | </p> | ||
</div> | </div> | ||
<div> | <div> | ||
| − | <h2 id="P1">II. Period 1 - | + | <h2 id="P1">II. Period 1 - CsgA - SpyTag</h2> |
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
| − | <div style="width: | + | <p class="text"> |
| + | We appended peptide domain SpyTag to amyloid protein CsgA, the dominant component in E. coli biofilms [Fig 1.1]. The sequence of this fusion protein was constructed on pET28a plasmid and was promoted by a Ptac promoter. Since there is already a copy of CsgA gene on MG1655 wild type’s genome, we need to knock out this wildtype gene CsgA. Using CRISPR, gene CgsA successfully knocked out the gene on MG1655’s genome (ΔCsgA-MG1655 strain, done by Yiming Dong from Bluepha). The strain ΔCsgA-MG1655 would then be used as chassis cell in afterward experiments. | ||
| + | </p> | ||
| + | <div style="width:46vw" class="content_pic_centre"> | ||
<img class="pictures" id = "21000" src="https://static.igem.org/mediawiki/2018/b/b9/T--SHSBNU_China--21000.png"/> | <img class="pictures" id = "21000" src="https://static.igem.org/mediawiki/2018/b/b9/T--SHSBNU_China--21000.png"/> | ||
| − | <p class="pic_text"></p> | + | <p class="pic_text">Fig1.1: Assembly and function of CsgA-SpyTag</p> |
</div> | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | + | <strong>Verification of biofilm forming</strong> | |
</p> | </p> | ||
| − | <div style="width: | + | <p class="text"> |
| + | Amyloid protein, like CsgA, could be colored by certain protein pigment. For instance, brilliant blue and Congo red. To verify the biofilm forming after IPTG induction, we compared the biofilm configuration between different experiment groups. | ||
| + | </p> | ||
| + | <p class="text"> | ||
| + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#APV">Protocol for amyloid protein verification</a> | ||
| + | </p> | ||
| + | <div style="width:46vw" class="content_pic_centre"> | ||
| + | <img class="pictures" id = "21003" src="https://static.igem.org/mediawiki/2018/1/12/T--SHSBNU_China--21003.png"style="width:100%"/> | ||
| + | <p class="pic_text">Fig 1.2: We set MG1655 Wild Type as positive control group, ΔCsgA-MG1655 with pET28a plasmid as negative control group, ΔCsgA-MG1655 with Ptac promoter-CsgA-SpyTag as experiment group.</p> | ||
| + | </div> | ||
| + | <p class="text"> | ||
| + | We discover [Fig1.2] that colonies of CsgA-SpyTag and the positive group show similar color and spreading pattern, while the colonies of the negative control group was colored to dark purple. Thus, our CsgA-SpyTag amyloid protein was successfully expressed. | ||
| + | </p> | ||
| + | <p class="text"> | ||
| + | We designed another experiments to measure the capture ability of CsgA–SpyTag [Fig 1.3]. We used sfGFP – SpyCatcher protein as the indicator [Fig 1.4]. Gene CsgA on the plasmid of pET28a was transferred in to ΔCsgA-MG1655 as control group. Gene CsgA – Spytag on the plasmid of pET28a was transferred in to ΔCsgA-MG1655 as experimental group. We used a sfGFP – SpyCatcher protein containing reaction stock, of which the fluorescence is measured, to react with the palette of both groups for 1 hour. Then, we centrifuged the reaction system and measure the fluorescence of the supernatant. The decrease of florescence indicated the amount of sfGFP – SpyCatcher protein the biofilm captured. | ||
| + | </p> | ||
| + | <p class="text"> | ||
| + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#SSV">Protocol for SpyTag-SpyCatcher system verification</a> | ||
| + | </p> | ||
| + | <div style="overflow:hidden"> | ||
| + | <div style="width:26vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "21004" src="https://static.igem.org/mediawiki/2018/7/74/T--SHSBNU_China--21004.png"/> | ||
| + | <p class="pic_text">Fig 1.3: Diagram of SpyTag-SpyCatcher system verification</p> | ||
| + | </div> | ||
| + | <div style="width:16vw" class="content_pic_right"> | ||
<img class="pictures" id = "21001" src="https://static.igem.org/mediawiki/2018/8/87/T--SHSBNU_China--21001.jpg"/> | <img class="pictures" id = "21001" src="https://static.igem.org/mediawiki/2018/8/87/T--SHSBNU_China--21001.jpg"/> | ||
| − | <p class="pic_text">Reaction stock leftover in experiment</p> | + | <p class="pic_text">Fig 1.4: Reaction stock leftover in experiment</p> |
</div> | </div> | ||
| + | <div style="width:46vw" class="content_pic_right"> | ||
| + | <img class="pictures" id = "21005" src="https://static.igem.org/mediawiki/2018/0/0e/T--SHSBNU_China--21005.png"/> | ||
| + | <p class="pic_text">Fig 1.5: Florescence measurement. Group1: MG1655 wildtype with no vector gene. Group2: ΔCsgA-MG1655 with Ptac promoter-CsgA. Group3: ΔCsgA-MG1655 with Ptac Promoter-CsgA-SpyTag.</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
<p class="text"> | <p class="text"> | ||
| − | + | As can be seen from the result, the experiment group showed the most decrease of sfGFP-SpyCatcher protein. The difference between the control group and experiment group is significant. Thus we can confirm our CsgA-SpyTag system is functional. | |
</p> | </p> | ||
| − | <div style="width: | + | <p class="text"> |
| − | <img class="pictures" id = " | + | After that, we test our biofilm’s ability to attach to surfaces for our hardware design. We induce ΔCsga-MG1655 with Ptac promoter-CsgA-SpyTag while PHA plastic beads were also soaked in the culture [Fig 1.6]. The PHA beads after 20 hours of induction was set as experiment group. Untreated, beads were set as control group. |
| − | <p class="pic_text"></p> | + | </p> |
| + | |||
| + | <div style="width:46vw" class="content_pic_right"> | ||
| + | <img class="pictures" id = "21006" src="https://static.igem.org/mediawiki/2018/6/6c/T--SHSBNU_China--21006.png"/> | ||
| + | <p class="pic_text">Fig 1.6: PHA plastic beads in culture</p> | ||
</div> | </div> | ||
| + | |||
<p class="text"> | <p class="text"> | ||
| − | + | We then use brilliant blue G250 to dye the both the experiment group and the control group. We allow 5 minute before washing off the extra dye with water for several time. | |
</p> | </p> | ||
| + | |||
| + | <div style="width:46vw" class="content_pic_right"> | ||
| + | <img class="pictures" id = "21007" src="https://static.igem.org/mediawiki/2018/7/75/T--SHSBNU_China--21007.png"/> | ||
| + | <p class="pic_text">Fig 1.7: Photos of the beginning and the end of the experiment.</p> | ||
| + | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | + | There is a visual difference [Fig 1.7] between the control and the experiment group, whit the experiment slightly more blue that the control group. | |
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | + | Though having a naked-eye distinguishable difference group between the experiment and control group, the difference was limited. In other words, it means our biofilm’s ability to stick to untreated PHA plastic beads is limited. To stick our biofilm on those beads, we need certain modifications like carving drills on PHA plastic beads. | |
</p> | </p> | ||
</div> | </div> | ||
<div> | <div> | ||
| − | <h2 id="P2">III. Period 2 - | + | <h2 id="P2">III. Period 2 - CotA - SpyCatcher</h2> |
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
<p class="text"> | <p class="text"> | ||
| − | Gene | + | Gene CotA from B. subtilis could translate as laccase in E. coli cells [Fig 2.1]. The gene was cloned on to pET28a plasmid, promoted by T7 promoter and could be induced by IPTG. |
</p> | </p> | ||
| − | <div style="width: | + | <div style="width:46vw" class="content_pic_left"> |
| − | <img class="pictures" id = " | + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/5/5e/T--SHSBNU_China--22010.png"/> |
| − | <p class="pic_text"></p> | + | <p class="pic_text">Fig 2.1: CotA promoted by T7 promoter.</p> |
</div> | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | We then added the | + | We then added the SpyCatcher sequence ahead of CotA gene [Fig 2.2] so that we can produce laccase with spycatcher. |
</p> | </p> | ||
| − | <div style="width: | + | <div style="width:46vw" class="content_pic_left"> |
| − | <img class="pictures" id = " | + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/c/c3/T--SHSBNU_China--22011.png"/> |
| − | <p class="pic_text"></p> | + | <p class="pic_text">Fig 2.2: CotA-SpyCatcher promoted by T7 promoter.</p> |
</div> | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | We also figured a way to secret our CotA protein out of the host cell. Signal peptides are | + | We also figured a way to secret our CotA protein out of the host cell [Fig 2.3]. Signal peptides are capable to lead the behind it out of the cell. A signal peptide sequence was also added ahead of cotA sequence. Three signal peptides was tested: Pelb, PhoA, OmpA. |
</p> | </p> | ||
| − | <div style="width: | + | <div style="width:46vw" class="content_pic_left"> |
| − | <img class="pictures" id = " | + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/4/43/T--SHSBNU_China--22012.png"/> |
| − | <p class="pic_text"></p> | + | <p class="pic_text">Fig 2.3: Signal peptides-CotA</p> |
</div> | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | + | We first tried to visualize the oxidation function of the laccase. So we made several solid LB media with Azo dyes mix in them and try to let bacteria decompose the pigment [Fig 2.4]. However, our laccase activity doesn’t seem to be better than the negative control group. Therefore, we need to find a quantitative method to measure laccase activity. | |
</p> | </p> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/1/15/T--SHSBNU_China--22013.jpeg"/> | ||
| + | <p class="pic_text">Fig2.4: Azo dyes mixed with solid LB media.</p> | ||
| + | </div> | ||
<p class="text"> | <p class="text"> | ||
| − | + | ABTS is a substance that is wildly used in commercial laccase activity assay kits. ABTS is in a clear solution and when it reacts with laccase, it would turn to a bullish-green color that has an absorbance peak at 420nm. We used laccase activity assay kit from Solarbio, type BC1635 which functions based on the previous mentioned method. We set a negative control group by transferring pET28a plasmid in to BL21 – DE3 strain. We set 4 experiment groups by transferring cotA, OmpA - cotA, PhoA - cotA, PelB – cotA gene into BL21 – DE3. We measured the laccase activity of all 5 groups mentioned above to check if our laccase is functional [Fig 2.5] [Fig 2.6] [Fig 2.7]. | |
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#LAA">Protocol for laccase activity assay, scale of protein</a> | |
| + | </p> | ||
| + | <p class="text"> | ||
| + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#LAM">Protocol for Laccase activity measuring kit</a | ||
| + | </p> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/4/42/T--SHSBNU_China--22014.png"/> | ||
| + | <p class="pic_text">Fig 2.5: ABTS measurement of laccase activity</p> | ||
| + | </div> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/4/41/T--SHSBNU_China--22015.png"/> | ||
| + | <p class="pic_text">Fig 2.6:Experiment diagram</p> | ||
| + | </div> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/c/cc/T--SHSBNU_China--22016.png"/> | ||
| + | <p class="pic_text">Fig 2.7: Result of ABTS laccase activity measurement, activity is calculated using the “cell count formula” in protocol</p> | ||
| + | </div> | ||
| + | <p class="text"> | ||
| + | From the laccase activity, we can see that CotA and Pelb-CotA turned out to be functional. However not all signal peptide sequence is functional. | ||
</p> | </p> | ||
</div> | </div> | ||
<div> | <div> | ||
| − | <h2 id="P3">IV. Period 3 - | + | <h2 id="P3">IV. Period 3 - Biofilm x Laccase</h2> |
</div> | </div> | ||
<div class="content"> | <div class="content"> | ||
<p class="text"> | <p class="text"> | ||
| − | The final | + | The final section is to combine laccase and biofilm together and test the laccase activity of Biofilm x Laccase system as a whole. Four group of experiments was conducted. |
</p> | </p> | ||
| − | <table bgcolor="#f0f0f0" cellspacing="1px"> | + | <div style="width:auto;overflow: hidden" class="content_pic_left"> |
| + | <table bgcolor="#f0f0f0" cellspacing="1px"> | ||
| + | <tr bgcolor="#f0f0f0"> | ||
| + | <td>Group name (in protocol, step 13)</td> | ||
| + | <td>Biofilm + SypTag</td> | ||
| + | <td>Enzyme</td> | ||
| + | </tr> | ||
| − | + | <tr bgcolor="#f0f0f0"> | |
| − | + | <td>1</td> | |
| − | + | <td>No</td> | |
| − | + | <td>CotA</td> | |
| − | + | </tr> | |
| − | + | <tr bgcolor="#f0f0f0"> | |
| − | + | <td>2</td> | |
| − | + | <td>No</td> | |
| − | + | <td>CotA - SpyCatcher</td> | |
| − | + | </tr> | |
| − | + | <tr bgcolor="#f0f0f0"> | |
| − | + | <td>3</td> | |
| − | + | <td>Yes</td> | |
| − | + | <td>CotA</td> | |
| − | + | </tr> | |
| − | + | <tr bgcolor="#f0f0f0"> | |
| − | + | <td>4</td> | |
| − | + | <td>Yes</td> | |
| − | + | <td>CotA - SpyCatcher</td> | |
| − | + | </tr> | |
| − | + | </table> | |
| − | < | + | <p class="pic_text">Table 3.1: Experiment form [fig 3.4]</p> |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
<p class="text"> | <p class="text"> | ||
The method of centrifuging and removing the supernatant was to remove uncombined enzyme after the reaction of SpyTag and SpyCatcher (Step 14). | The method of centrifuging and removing the supernatant was to remove uncombined enzyme after the reaction of SpyTag and SpyCatcher (Step 14). | ||
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#BXL">Protocol for Biofilm x laccase</a> | |
</p> | </p> | ||
<p class="text"> | <p class="text"> | ||
| − | + | <a href="https://2018.igem.org/Team:SHSBNU_China/Protocol#LAM">Protocol for Laccase activity measurement kit</a> | |
</p> | </p> | ||
| − | <div style="width: | + | <div style="width:46vw" class="content_pic_left"> |
| − | <img class="pictures" id = " | + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/4/46/T--SHSBNU_China--23010.jpg"/> |
| − | <p class="pic_text"></p> | + | <p class="pic_text">Fig3.1: Diagram of experiment.</p> |
</div> | </div> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/d/d5/T--SHSBNU_China--23011.jpg"/> | ||
| + | <p class="pic_text">Fig 3.2: Laccase activity assay result of supernatant</p> | ||
| + | </div> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/a/af/T--SHSBNU_China--23012.jpg"/> | ||
| + | <p class="pic_text">Fig3.3: laccase activity assay result of lysate</p> | ||
| + | </div> | ||
| + | <div style="width:46vw" class="content_pic_left"> | ||
| + | <img class="pictures" id = "" src="https://static.igem.org/mediawiki/2018/4/4a/T--SHSBNU_China--23000.jpg"/> | ||
| + | <p class="pic_text">Fig 3.4: Laccase activity assay of the Biofilm x Laccase system as a whole.</p> | ||
| + | </div> | ||
| + | <p class="text"> | ||
| + | The measurement result indicated by figure 3.2 and 3.3 suggests that SpyCatcher-CotA is more concentrated in the lysate rather than supernatant. The * notation stand for the significance of difference in a group, the more * a group has the more significant the difference in that group would be. Significant difference only occurs when SpyCatcher-CotA was used to fix onto the biofilm or the negative control group. Which means that SpyCatcher-CotA can combines effectively with CsgA-Spytag. Till now, all the basic concept of Biofilm x Laccase is proven to be feasible and functional in lab conditions. | ||
| + | </p> | ||
</div> | </div> | ||
Latest revision as of 03:56, 18 October 2018

Experiment
I. Outline
Our system includes two main parts one is CotA laccase and the other is biofilm containing SpyTag. So our experiments were mainly divided in three periods, verifying both parts individually and verifying Biofilm x Laccase system as a whole.
The first period was to construct our biofilm containing SpyTag. We fused SpyTag to the CsgA, which is the major component of our biofilm. We successfully verified that protein fused with spycatcher domain could be fixed on our biofilm.
Then we isolated CotA gene of B. subtilis WT168 and successfully expressed this laccase in E. coli cell. We confirmed its oxidation function. Furthermore, we attached CotA coding sequence with several signal peptides and tested the efficiency of secretion.
The final period is mainly focused on combination of the two parts above. Experiments were conducted to measure the laccase activity of the total Biofilm x Laccase system. A significant difference was confirmed between the experiment group and the control group. Thus, our system is successful.
II. Period 1 - CsgA - SpyTag
We appended peptide domain SpyTag to amyloid protein CsgA, the dominant component in E. coli biofilms [Fig 1.1]. The sequence of this fusion protein was constructed on pET28a plasmid and was promoted by a Ptac promoter. Since there is already a copy of CsgA gene on MG1655 wild type’s genome, we need to knock out this wildtype gene CsgA. Using CRISPR, gene CgsA successfully knocked out the gene on MG1655’s genome (ΔCsgA-MG1655 strain, done by Yiming Dong from Bluepha). The strain ΔCsgA-MG1655 would then be used as chassis cell in afterward experiments.

Fig1.1: Assembly and function of CsgA-SpyTag
Verification of biofilm forming
Amyloid protein, like CsgA, could be colored by certain protein pigment. For instance, brilliant blue and Congo red. To verify the biofilm forming after IPTG induction, we compared the biofilm configuration between different experiment groups.
Protocol for amyloid protein verification
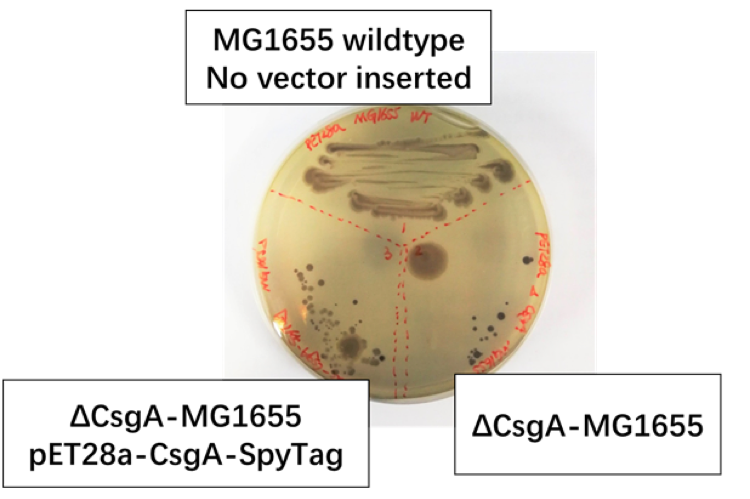
Fig 1.2: We set MG1655 Wild Type as positive control group, ΔCsgA-MG1655 with pET28a plasmid as negative control group, ΔCsgA-MG1655 with Ptac promoter-CsgA-SpyTag as experiment group.
We discover [Fig1.2] that colonies of CsgA-SpyTag and the positive group show similar color and spreading pattern, while the colonies of the negative control group was colored to dark purple. Thus, our CsgA-SpyTag amyloid protein was successfully expressed.
We designed another experiments to measure the capture ability of CsgA–SpyTag [Fig 1.3]. We used sfGFP – SpyCatcher protein as the indicator [Fig 1.4]. Gene CsgA on the plasmid of pET28a was transferred in to ΔCsgA-MG1655 as control group. Gene CsgA – Spytag on the plasmid of pET28a was transferred in to ΔCsgA-MG1655 as experimental group. We used a sfGFP – SpyCatcher protein containing reaction stock, of which the fluorescence is measured, to react with the palette of both groups for 1 hour. Then, we centrifuged the reaction system and measure the fluorescence of the supernatant. The decrease of florescence indicated the amount of sfGFP – SpyCatcher protein the biofilm captured.
Protocol for SpyTag-SpyCatcher system verification
As can be seen from the result, the experiment group showed the most decrease of sfGFP-SpyCatcher protein. The difference between the control group and experiment group is significant. Thus we can confirm our CsgA-SpyTag system is functional.
After that, we test our biofilm’s ability to attach to surfaces for our hardware design. We induce ΔCsga-MG1655 with Ptac promoter-CsgA-SpyTag while PHA plastic beads were also soaked in the culture [Fig 1.6]. The PHA beads after 20 hours of induction was set as experiment group. Untreated, beads were set as control group.

Fig 1.6: PHA plastic beads in culture
We then use brilliant blue G250 to dye the both the experiment group and the control group. We allow 5 minute before washing off the extra dye with water for several time.

Fig 1.7: Photos of the beginning and the end of the experiment.
There is a visual difference [Fig 1.7] between the control and the experiment group, whit the experiment slightly more blue that the control group.
Though having a naked-eye distinguishable difference group between the experiment and control group, the difference was limited. In other words, it means our biofilm’s ability to stick to untreated PHA plastic beads is limited. To stick our biofilm on those beads, we need certain modifications like carving drills on PHA plastic beads.
III. Period 2 - CotA - SpyCatcher
Gene CotA from B. subtilis could translate as laccase in E. coli cells [Fig 2.1]. The gene was cloned on to pET28a plasmid, promoted by T7 promoter and could be induced by IPTG.

Fig 2.1: CotA promoted by T7 promoter.
We then added the SpyCatcher sequence ahead of CotA gene [Fig 2.2] so that we can produce laccase with spycatcher.

Fig 2.2: CotA-SpyCatcher promoted by T7 promoter.
We also figured a way to secret our CotA protein out of the host cell [Fig 2.3]. Signal peptides are capable to lead the behind it out of the cell. A signal peptide sequence was also added ahead of cotA sequence. Three signal peptides was tested: Pelb, PhoA, OmpA.

Fig 2.3: Signal peptides-CotA
We first tried to visualize the oxidation function of the laccase. So we made several solid LB media with Azo dyes mix in them and try to let bacteria decompose the pigment [Fig 2.4]. However, our laccase activity doesn’t seem to be better than the negative control group. Therefore, we need to find a quantitative method to measure laccase activity.

Fig2.4: Azo dyes mixed with solid LB media.
ABTS is a substance that is wildly used in commercial laccase activity assay kits. ABTS is in a clear solution and when it reacts with laccase, it would turn to a bullish-green color that has an absorbance peak at 420nm. We used laccase activity assay kit from Solarbio, type BC1635 which functions based on the previous mentioned method. We set a negative control group by transferring pET28a plasmid in to BL21 – DE3 strain. We set 4 experiment groups by transferring cotA, OmpA - cotA, PhoA - cotA, PelB – cotA gene into BL21 – DE3. We measured the laccase activity of all 5 groups mentioned above to check if our laccase is functional [Fig 2.5] [Fig 2.6] [Fig 2.7].
Protocol for laccase activity assay, scale of protein
Protocol for Laccase activity measuring kit

Fig 2.5: ABTS measurement of laccase activity

Fig 2.6:Experiment diagram

Fig 2.7: Result of ABTS laccase activity measurement, activity is calculated using the “cell count formula” in protocol
From the laccase activity, we can see that CotA and Pelb-CotA turned out to be functional. However not all signal peptide sequence is functional.
IV. Period 3 - Biofilm x Laccase
The final section is to combine laccase and biofilm together and test the laccase activity of Biofilm x Laccase system as a whole. Four group of experiments was conducted.
The method of centrifuging and removing the supernatant was to remove uncombined enzyme after the reaction of SpyTag and SpyCatcher (Step 14).
Protocol for Biofilm x laccase
Protocol for Laccase activity measurement kit

Fig3.1: Diagram of experiment.

Fig 3.2: Laccase activity assay result of supernatant

Fig3.3: laccase activity assay result of lysate

Fig 3.4: Laccase activity assay of the Biofilm x Laccase system as a whole.
The measurement result indicated by figure 3.2 and 3.3 suggests that SpyCatcher-CotA is more concentrated in the lysate rather than supernatant. The * notation stand for the significance of difference in a group, the more * a group has the more significant the difference in that group would be. Significant difference only occurs when SpyCatcher-CotA was used to fix onto the biofilm or the negative control group. Which means that SpyCatcher-CotA can combines effectively with CsgA-Spytag. Till now, all the basic concept of Biofilm x Laccase is proven to be feasible and functional in lab conditions.





