| Line 72: | Line 72: | ||
| − | <body class="stretched"> | + | <body class="stretched sticky-responsive-pagemenu"> |
<!-- Document Wrapper | <!-- Document Wrapper | ||
| Line 162: | Line 162: | ||
============================================= --> | ============================================= --> | ||
| − | <section | + | <section style="overflow:hidden; position:relative; background-color: #FFF;"> |
<div class="content-wrap"> | <div class="content-wrap"> | ||
| Line 231: | Line 231: | ||
<p style="margin-bottom: 0in; line-height: 300%"><br/></p> | <p style="margin-bottom: 0in; line-height: 300%"><br/></p> | ||
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/7/7a/T--Vilnius-Lithuania-OG--fig1a.png" name="fig1a"></div> |
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/6/66/T--Vilnius-Lithuania-OG--fig1b.png" name="fig1b"></div> |
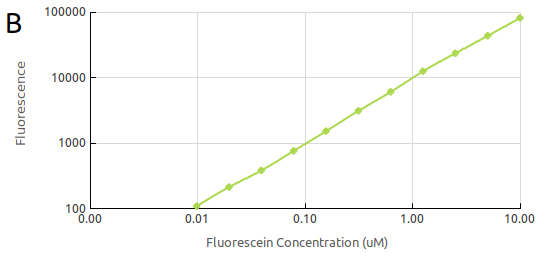
<p style="margin-bottom: 0in; line-height: 200%"><center> <b> Figure | <p style="margin-bottom: 0in; line-height: 200%"><center> <b> Figure | ||
| Line 255: | Line 255: | ||
<p style="margin-bottom: 0in; line-height: 300%"><br/></p> | <p style="margin-bottom: 0in; line-height: 300%"><br/></p> | ||
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/0/01/T--Vilnius-Lithuania-OG--fig2a.png" name="fig1a"></div> |
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/8/8c/T--Vilnius-Lithuania-OG--fig2b.png" name="fig1b"></div> |
| Line 299: | Line 299: | ||
<p style="margin-bottom: 0in; line-height: 300%"><br/></p> | <p style="margin-bottom: 0in; line-height: 300%"><br/></p> | ||
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/7/7d/T--Vilnius-Lithuania-OG--fig3a.png" name="fig3a"></div> |
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/b/b4/T--Vilnius-Lithuania-OG--fig3b.png" name="fig3b"></div> |
<p style="margin-bottom: 0in; line-height: 200%"><br/></p> | <p style="margin-bottom: 0in; line-height: 200%"><br/></p> | ||
| Line 329: | Line 329: | ||
<p style="margin-bottom: 0in; line-height: 300%"><br/></p> | <p style="margin-bottom: 0in; line-height: 300%"><br/></p> | ||
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/7/76/T--Vilnius-Lithuania-OG--fig4a.png" name="fig3a"></div> |
| − | <div align="center"><img src="/ | + | <div align="center"><img src="https://static.igem.org/mediawiki/2018/4/41/T--Vilnius-Lithuania-OG--fig4b.png" name="fig3b"></div> |
<p style="margin-bottom: 0in; line-height: 200%"><br/></p> | <p style="margin-bottom: 0in; line-height: 200%"><br/></p> | ||
Revision as of 04:07, 17 October 2018