Methods and Results
1. Calibration protocols
At
firstly, we measured absorbance (Abs600)
of weakly scattering LUDOX CL-X to obtain a multiplication factor of
Abs600
to transform absorbance values to the comparable OD600
units.
Following
that, we prepared serial dilutions of monodisperse silica
microspheres and obtained standard particle curve (Fig.
1) which allows to
convert Abs600 measurements to
an estimated number of cells.
Figure
1. Standard particle curve obtained by using microspheres that
are similar size to a cell displayed in (A) a linear scale and
(B) a log scale.
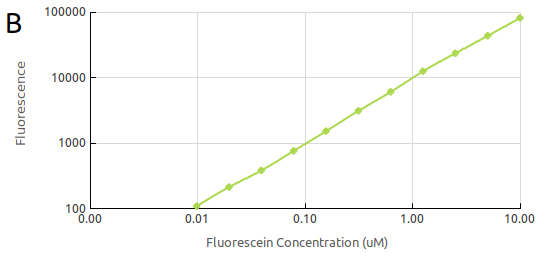
The
final calibration was that of fluorescein concentration. We generated
a standard curve of fluorescence for fluorescein concentration by,
again, making serial dilutions of fluorescein (Fig.
2). This way we
could convert our cell based readings of fluorescence to an
equivalent fluorescein concentration. The measurements were taken
with the same setting as those which would be used measuring cell
fluorescence.
Figure 2.
Fluorescein standard curve obtained by serial dilutions in (A)
the linear scale and in (B) the log scale.
2. Cell measurement protocol
We
were provided with four different devices (Test device 1 -
BBa_J364000,
Test device 2 - BBa_J364001,
Test device 3 - BBa_J364002,
Test device 4 - BBa_J364007,
Test device 5 - BBa_J364008,
Test device 6 - BBa_J364009)
along with negative (BBa_R0040)
and positive
(BBa_I20270)
controls which then we transformed into E.
coli DH5α
strain. Devices express GFP under Anderson promoters of different
strength and are in the pSB1C3 backbone which carries resistance to
chloramphenicol.
We
made overnight cultures of two colonies of each transformation plate.
The next day, the cultures were diluted to OD600=0.02
and grown for 6 hours. The absorbance (600 nm) (Fig. 3A) and
fluorescence (485/520, gain = 50) (Fig. 3B) of cultures were measured
at 0 and 6 hours.
Figure 3.
Average absorbance (A) and fluorescence (B) measurements of the 2 colonies and 4 replicates for each
sample, taken after 6 hours of growth.
The
negative control demonstrates the highest absorbance which could be
the result of the lowest metabolic load of all the devices, since it
has no GFP expressed. On the other hand, devices 1, 4 and 5 show much
slower bacteria growth even though the fluorescence of these devices
is not the highest. Furthermore, device 1 demonstrates a really
moderate absorbance and fluorescence increase after 6 hours
suggesting that environmental factors could have impeded sample
growth.
Next,
the previous data (absorbance and fluorescence) was normalized to the
comparable OD units (Fig. 4A) and then to particles (Fig. 4B) so that we could
determine the mean expression level of GFP per cell.
Figure 4.
Normalization of fluorescence to the OD units (A) and MFEL per particle (B).
After
6 hours all the devices (except the negative control) demonstrates a
relatively decreased level of fluorescence. That could be explained
by cells reaching a certain level of GFP expression after which the
fluorescence stops increasing whereas absorption steadily goes up.
3. Colony forming units (CFU) per 0.1 OD600
The
second approach is to calibrate OD600
to CFU which reflects the cell count in the sample. For this approach
only negative and positive controls were used.
The
overnight cultures of two colonies per control were diluted to
OD600=0.1
(three replications) and serial dilutions were made which then were
spreaded on agar plates. The next morning the colonies were counted
to find out the number of cells in the samples (Fig. 5).
Figure 5.
Normalization of fluorescence to the OD units (A) and MFEL per particle (B).
NC
1.1 and PC 1.2 samples show relatively high and low values
resspectively comparing to other samples which could indicate a
systematic error in a serial dilution step.
Conclusions
In this study cell fluorescence depends on the strength of the promotor under which GFP was expressed. However, all the cells with different devices demonstrated a reduced fluorescence per cell after 6 hours indicating a slowed down GFP production.