|
|
| Line 4: |
Line 4: |
| | <html> | | <html> |
| | <head> | | <head> |
| − | <script>
| |
| − | </script>
| |
| − | <style>
| |
| | | | |
| − |
| |
| − | </style>
| |
| − |
| |
| − |
| |
| − | </head>
| |
| − |
| |
| − | <body>
| |
| − | <div class="container">
| |
| − | <div class="content">
| |
| − | <div class="titleimg">
| |
| − | <h1>Modelling</h1>
| |
| − | <br/>
| |
| − | <br/>
| |
| − | <img src="https://static.igem.org/mediawiki/2018/5/59/T--Imperial_College--modelling.png" alt="" width="24%"; >
| |
| − | </div>
| |
| − |
| |
| − | <div class="what">
| |
| − | <h3>What is Modelling?</h3>
| |
| − | <p2>Constructing and manipulating abstract (mathematical and/or graphical) representations of economic, engineering, manufacturing, social, and other types of situations and natural phenomenon, simulated with the help of a computer system. Also called computer simulation.
| |
| − | </p2>
| |
| − | <br/>
| |
| − | <br/>
| |
| − | </div>
| |
| − | <div class="why">
| |
| − | <h3>Why is Modelling important in biological studies?</h3>
| |
| − | <p2>Computer modeling simulates the interaction among many different variables to provide scientists with a general idea of how a biological system may work.</p2>
| |
| − | </div>
| |
| − |
| |
| − |
| |
| − | <div class="accordion">
| |
| − | <h3>Our Computer Models</h3>
| |
| − |
| |
| − | <button class="collapsible" type="button">Electrochemical Model</button>
| |
| − | <div class="drop">
| |
| | <style> | | <style> |
| | | | |
| − | #expt1, #expt2, #expt3, #expt4, #expt5, #expt6,#expt7,#expt8,#expt9 { | + | #expt1, #expt2, #expt3, #expt4, #expt5, #expt6,#expt7,#expt8,#expt9 { |
| | width:100%; | | width:100%; |
| | height:5px; | | height:5px; |
| | margin-bottom:6%; | | margin-bottom:6%; |
| | } | | } |
| − |
| |
| − | .contents ul{
| |
| − | margin-top:6%;
| |
| − | box-sizing: border-box;
| |
| − | margin-left:10%;
| |
| − | list-style-type: none;
| |
| − | background:none;
| |
| − | }
| |
| − |
| |
| − | .contents{
| |
| − | margin:auto;
| |
| − | margin-left:30%;
| |
| − | width: 50%;
| |
| − | }
| |
| − |
| |
| − | .contents ul li{
| |
| − | width: 100%;
| |
| − | margin-bottom:5%;
| |
| − | cursor: pointer;
| |
| − | border-radius:20px;
| |
| − | background:none;
| |
| − | transition: border 0.3s ease;
| |
| − | box-sizing:border-box;
| |
| − | }
| |
| − |
| |
| − | .contents ul li a{
| |
| − | text-decoration: none;
| |
| − | font-size: 25px;
| |
| − | font-family: 'Varela Round', sans-serif;
| |
| − | color: #374785;
| |
| − | }
| |
| − |
| |
| − | .contents ul li a:hover{
| |
| − | font-weight:bold;
| |
| − | }
| |
| − |
| |
| | | | |
| | | | |
| Line 163: |
Line 90: |
| | box-sizing: border-box; | | box-sizing: border-box; |
| | } | | } |
| − | .tg {border-collapse:collapse;border-spacing:0;} | + | .caption p{ |
| − | .tg td{font-family:Arial, sans-serif;font-size:14px;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:black;}
| + | margin:auto !important; |
| − | .tg th{font-family:Arial, sans-serif;font-size:14px;font-weight:normal;padding:10px 5px;border-style:solid;border-width:1px;overflow:hidden;word-break:normal;border-color:black;}
| + | text-align: center !important; |
| − | .tg .tg-1wig{font-weight:bold;text-align:left;vertical-align:top}
| + | |
| − | .tg .tg-096r{color:#000000;text-align:left;vertical-align:top}
| + | |
| − | .tg .tg-0lax{text-align:left;vertical-align:top}
| + | |
| − | </style>
| + | |
| | | | |
| | + | } |
| | + | </style> |
| | </head> | | </head> |
| | | | |
| − | <body>
| |
| − | <div class="container">
| |
| − | <div class="content">
| |
| | | | |
| − | <div class="titleimg">
| + | <body> |
| − | <h1>Methods</h1>
| + | <div class="container"> |
| − | <br/>
| + | <div class="content"> |
| − | <br/>
| + | <div class="titleimg"> |
| − | <img src="https://static.igem.org/mediawiki/2018/7/71/T--Imperial_College--experiment.png" alt="" width="28%"; >
| + | <h1>Modelling</h1> |
| − | </div>
| + | <br/> |
| | + | <br/> |
| | + | <img src="https://static.igem.org/mediawiki/2018/5/59/T--Imperial_College--modelling.png" alt="" width="24%"; > |
| | + | </div> |
| | | | |
| − | <h3>Contents</h3>
| + | <!--Start Experiment 1--> |
| − |
| + | |
| − | <div class="contents">
| + | |
| − | <ul>
| + | |
| − | <li><a href="#expt1">Assembling the Pixcell Constructs</a></li>
| + | |
| − | <li><a href="#expt2">Characterizing Pixcell Constructs</a></li>
| + | |
| − | <li><a href="#expt3">Electrochemistry of Redox Modulators</a></li>
| + | |
| − | <li><a href="#expt4">Spatial Electronic Control of Gene Induction</a></li>
| + | |
| − | <li><a href="#expt5">Creating the Sox Library</a></li>
| + | |
| − | <li><a href="#expt6">Characterising the Sox Library</a></li>
| + | |
| − | <li><a href="#expt7">Testing Phenazine Methosulfate</a></li>
| + | |
| − | <li><a href="#expt8">Biocontainment- Gp2 Growth Inhibition</a></li>
| + | |
| − | <li><a href="#expt9">Fabric Bioprinting - Melanin</a></li>
| + | |
| − | | + | |
| − | </ul>
| + | |
| − | </div>
| + | |
| − | | + | |
| − |
| + | |
| − | | + | |
| − | <h3 class="marginbottom">Experiments</h3>
| + | |
| − | | + | |
| − |
| + | |
| − | <!--Start Experiment 1-->
| + | |
| | <div id="expt1"></div> | | <div id="expt1"></div> |
| − | <h4 class="underbold"> Assembling the Pixcell Constructs</h4> | + | <h4 class="underbold" > Assembling the Pixcell Constructs</h4> |
| | <div onclick="expandAll('expt1p');" class="expandButton"> | | <div onclick="expandAll('expt1p');" class="expandButton"> |
| − | <p class="expand expt1p colorChange" style="font-size:25px;padding-top:1px;" >Access All</p>
| + | <p class="expand expt1p colorChange" style="font-size:25px;padding-top:1px;" >Access All</p> |
| | <div class="expand" style="margin-right:5px;"> | | <div class="expand" style="margin-right:5px;"> |
| − | <i class="fa fa-angle-double-down expt1p"></i>
| + | <i class="fa fa-angle-double-down expt1p"></i> |
| | + | |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 220: |
Line 124: |
| | <div type="button" class="accordion"> Description </div> | | <div type="button" class="accordion"> Description </div> |
| | <div class="description"> | | <div class="description"> |
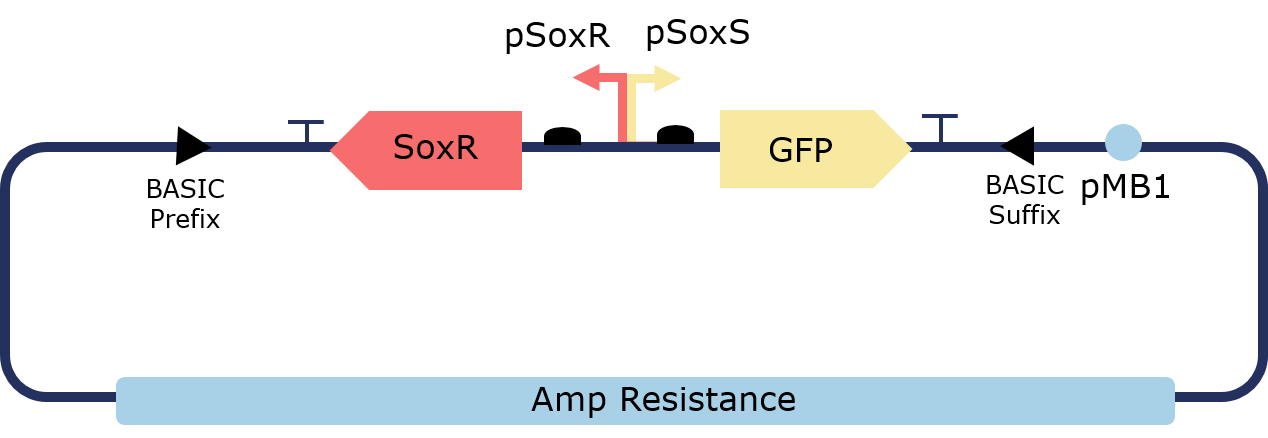
| − | <p>Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p> | + | <p> The PixCell Construct consists of a repurposed version of the soxRS regulon from E. coli, consisting of SoxR and GFP being expressed from either side of the pSoxR/pSoxS bidirectional promoter. pSoxR provides constitutive expression of SoxR. When oxidised, either directly by redox-cycling molecule or by oxidative stress, SoxR binds and activates transcription of GFP downstream of pSoxS.</p> |
| − | </div>
| + | <img class="center" src="https://static.igem.org/mediawiki/2018/6/6d/T--Imperial_College--PixcellConstructResults.png" alt="" width="50%";> |
| − | | + | |
| − | <div type="button" class="accordion">Materials</div>
| + | <div class="caption"> |
| − | <div class="panel expt1p">
| + | <p><b>Figure 1.</b> Illustration of the PixCell Construct</p> |
| − | <p >Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p>
| + | </div> |
| | + | |
| | + | |
| | + | |
| | + | </div> |
| | + | |
| | + | <div type="button" class="accordion">Relevance</div> |
| | + | <div class="panel expt1p"> |
| | + | |
| | + | <p> The Pixcell construct was designed to test whether we could control the expression of GFP by controlling the oxidation state of SoxR through our electrochemical set up.</p> |
| | </div> | | </div> |
| | | | |
| − | <div type="button" class="accordion">Notes</div>
| + | <div type="button" class="accordion">Results</div> |
| − | <div class="panel expt1p">
| + | <div class="panel expt1p" style="transition:max-height 1s ease-in-out;"> |
| − | <p>Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
| + | <p> Using the Golden Gate assembly method, with two inserts; the SOXRS regulon and GFP, which were originally amplified out from the E. coli genome and a storage vector respectively, the construct was assembled. Gel electrophoresis showed that a construct of approximately the right length was produced and the construct was then sequence verified. </p> |
| − | Lorem ipsum dolor, sit amet consectetur adipisicing elit. <a href="#" class="highlight">Competant cell protocol</a> Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p>
| + | <img class="figure" src="https://static.igem.org/mediawiki/2018/b/b3/T--Imperial_College--PixcellConstructGel.png"> |
| − | <p>Lorem ipsum dolor, sit amet <a href="#" class="highlight">Heatshock protocol</a> consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p>
| + | |
| − | </div>
| + | |
| | + | <div class="caption"> |
| | + | <p><b>Figure 1.</b> PCR results from the amplification of the entire construct</p> |
| | + | </div> |
| | + | |
| | + | <div class="clr"></div> |
| | + | |
| | + | <p>In order to lower the basal level of GFP expression a degradation tag was added. Therefore, any increase in GFP expression induced by the oxidation of SoxR would be more predominant.</p> |
| | + | |
| | + | <img class="figure" src="https://static.igem.org/mediawiki/2018/2/26/T--Imperial_College--PixcellConstructDEGTAG.png"> |
| | + | |
| | + | <div class="caption"> |
| | + | <p><b>Figure 1.</b> Illustration of the PixCell Construct DegTag</p> |
| | + | </div> |
| | + | |
| | + | </div> |
| | + | |
| | | | |
| | | | |
| | | | |
| − | <div type="button" class="accordion" >Procedure</div> | + | <div type="button" class="accordion" >Summary</div> |
| | <div class="panel expt1p"> | | <div class="panel expt1p"> |
| − | <p>Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p> | + | <p> The Pixcell construct enables us to test whether we can control gene expression by controlling the oxidation state of soxR. The Pixcell construct DegTag enables the system to be more dynamic in that seeing the system switch from on to off is quicker. </p> |
| | </div> | | </div> |
| | <div class="clr"></div> | | <div class="clr"></div> |
| Line 245: |
Line 174: |
| | | | |
| | | | |
| − | <!--Start Experiment 2-->
| + | <div class="what"> |
| − | <div id="expt2">
| + | <h3>What is Modelling?</h3> |
| − | <h4 class="underbold"> Characterizing Pixcell Constructs</h4>
| + | <p2>Constructing and manipulating abstract (mathematical and/or graphical) representations of economic, engineering, manufacturing, social, and other types of situations and natural phenomenon, simulated with the help of a computer system. Also called computer simulation. |
| − | <div onclick="expandAll('expt2p');" class="expandButton">
| + | </p2> |
| − | <p class="expand expt2p colorChange" style="font-size:25px;padding-top:1px;" >Access All</p>
| + | <br/> |
| − | <div class="expand" style="margin-right:5px;">
| + | <br/> |
| − | <i class="fa fa-angle-double-down expt2p"></i>
| + | </div> |
| − | </div>
| + | <div class="why"> |
| − | </div>
| + | <h3>Why is Modelling important in biological studies?</h3> |
| − | </div>
| + | <p2>Computer modeling simulates the interaction among many different variables to provide scientists with a general idea of how a biological system may work.</p2> |
| − | <div class="clr"></div>
| + | </div> |
| − |
| + | |
| − | <div type="button" class="accordion"> Description </div>
| + | |
| − | <div class="description">
| + | |
| − | <p>This protocol describes how to measure E. coli growth and GFP expression over time with a plate reader, using the optical density at 600 nm as signal for cell density and excitation and emission wavelengths of 475.
| + | |
| − | All characterization was performed using the FLUOstar BMG Labtech in greiner bioone F-bottom dark plates with clear bottom, to decrease fluorescence background.
| + | |
| − | Each cycle of this experiment takes two days and has to be repeated two days, in order to have 2 biological and 2 technical replicates, for a total of 4 days and 4 96-well plates used.
| + | |
| − | </p> | + | |
| − | </div>
| + | |
| − |
| + | |
| − | <div type="button" class="accordion">Materials</div>
| + | |
| − | <div class="panel expt2p">
| + | |
| − | <ul>
| + | |
| − | <li>Autoclaved LB media</li>
| + | |
| − | <li>Autoclaved 10x concentated LB media</li>
| + | |
| − | <li>500 uM pyocyanin stock solution (from Sigma-Aldrich, CAS: No. 86-66-5)</li>
| + | |
| − | <li>1250 mM potassium ferricyanide stock solution (from Sigma-Aldrich, CAS: No. 13746-66-2)</li>
| + | |
| − | <li>500 mM ferrocyanide stock solution (from Sigma-Aldrich, CAS 14459-95-1 )</li>
| + | |
| − | <li>2% Sodium Sulfite Solution</li>
| + | |
| − | <li>
| + | |
| − | Agar plates with colonies of E. coli DJ901 strains with respective constructs: PixCell Patterning Circuit 1 (BBa_K2862021). PixCell Patterning Circuit 2 (BBa_K2862022), negative control (DJ901 WT) and positive control (constitutively expressed GFP).</li>
| + | |
| − | <li>37°C shaking incubator</li>
| + | |
| − | <li>BMG Labtech FLUOstar plate reader</li>
| + | |
| − | <li>4x Greiner BioOne 96-F 96-well microplate (black with clear bottom)</li>
| + | |
| − | <li>14 mL culture tubes</li>
| + | |
| − | <li>autoclaved eppendorfs</li>
| + | |
| − | </ul>
| + | |
| − | </div>
| + | |
| | | | |
| − | <div type="button" class="accordion">Notes</div>
| + | <div class="accordion"> |
| − | <div class="panel expt2p">
| + | <h3>Our Computer Models</h3> |
| − | <p>Work in very sterile conditions. To prevent contaminations in the blanks, use triple antibiotics.
| + | |
| − | </br>
| + | <button class="collapsible" type="button">Electrochemical Model</button> |
| − | Store pyocyanin and ferrocyanide and ferricyanide solutions in the freezer until use.
| + | <div class="drop"> |
| − | </br>
| + | |
| − | We used 10x concentrated LB in order not to dilute nutrients at higher pyocyanin concentration; this was back diluted with autoclaved water. Mastemixes have double the working concentrations, as 100 uL of solution is then diluted with 100 uL of liquid cultures.
| + | |
| − | | + | |
| − | </p>
| + | |
| − | </div>
| + | |
| − |
| + | |
| − | <div type="button" class="accordion" >Procedure</div>
| + | |
| − | <div class="panel expt2p">
| + | |
| − | <p>Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.</p>
| + | |
| − | <ol>
| + | |
| − | <li>Grow starter liquid cultures of cells (Day 0, 10-15 min preparation + overnight)</li>
| + | |
| − | | + | |
| − | <p>Working in very sterile conditions, take 5 14mL culture tubes and prepare them according to table below: </p>
| + | |
| − | | + | |
| − | <table class="tg">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-1wig">Label</th> | + | |
| − | <th class="tg-1wig"><span style="font-style:italic">E. coli</span> strain</th> | + | |
| − | <th class="tg-1wig">Construct</th> | + | |
| − | <th class="tg-1wig">Antibiotic (Ab)</th>
| + | |
| − | <th class="tg-1wig">V Ab (uL)</th>
| + | |
| − | <th class="tg-1wig">V LB (mL)</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">PC1</td>
| + | |
| − | <td class="tg-0lax">DJ901</td>
| + | |
| − | <td class="tg-0lax">BBa_K2862021</td>
| + | |
| − | <td class="tg-096r">Kan + Amp</td>
| + | |
| − | <td class="tg-0lax">5 + 5</td>
| + | |
| − | <td class="tg-0lax">5</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">PC2</td>
| + | |
| − | <td class="tg-0lax">DJ901</td>
| + | |
| − | <td class="tg-0lax">BBa_K2862022</td>
| + | |
| − | <td class="tg-0lax">Kan + Amp</td>
| + | |
| − | <td class="tg-0lax">5 + 5</td>
| + | |
| − | <td class="tg-0lax">5</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">-</td>
| + | |
| − | <td class="tg-0lax">DJ901</td>
| + | |
| − | <td class="tg-0lax">none</td>
| + | |
| − | <td class="tg-0lax">Kan</td>
| + | |
| − | <td class="tg-0lax">5 </td>
| + | |
| − | <td class="tg-0lax">5</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">+</td>
| + | |
| − | <td class="tg-0lax">DJ901</td>
| + | |
| − | <td class="tg-0lax">GFP_Boo</td>
| + | |
| − | <td class="tg-0lax">Kan + Chl</td>
| + | |
| − | <td class="tg-0lax">5 + 5</td>
| + | |
| − | <td class="tg-0lax">5</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">Blanks</td>
| + | |
| − | <td class="tg-0lax">None</td>
| + | |
| − | <td class="tg-0lax">None</td>
| + | |
| − | <td class="tg-0lax">Kan + Amp + Chl</td>
| + | |
| − | <td class="tg-0lax">5 + 5</td>
| + | |
| − | <td class="tg-0lax">5</td>
| + | |
| − | </tr>
| + | |
| − | </table>
| + | |
| − | <p>With a pipette tip pick a desired colony* from an agar plate and release the contaminated tip in the culture tip. Place inoculated culture tubes in a 37° C in a shaking incubator overnight (for faster growth the angle between the vertical axis of the tube and the shaking plane of the incubator should be of 45°)</p>
| + | |
| − | | + | |
| − | <li>Prepare mastermixes (30 min)</li>
| + | |
| − | <p>Mastermixes for each redox molecule concentration are prepared in 1.5 mL eppendorfs. The mastermixes are enough to fill 2 96-well microplates with 100 uL of solutions in each well, in order to have two replicate of the same experiment. Take 8 autoclaved 1.5 mL eppendorfs and prepare them as described in the table below.</p>
| + | |
| − | <p>Mastermizes for Day 1 and Day 2 :</p>
| + | |
| − | <table class="tg">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-s268">Condition</th>
| + | |
| − | <th class="tg-s268">V Pyo (uL)</th>
| + | |
| − | <th class="tg-s268">Kan (uL)</th>
| + | |
| − | <th class="tg-0lax">LB 10x uL</th>
| + | |
| − | <th class="tg-0lax">Autoclaved Water (uL)</th>
| + | |
| − | <th class="tg-0lax">Tot Vol (uL)</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-s268">A = 0 uM Pyo</td>
| + | |
| − | <td class="tg-s268">0</td>
| + | |
| − | <td class="tg-s268">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1098</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-s268">B = 0.01 uM Pyo</td>
| + | |
| − | <td class="tg-s268">0.0976</td>
| + | |
| − | <td class="tg-s268">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1097.9024</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">C = 0.025 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">0.244</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1097.756</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">D = 0.05 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">0.488</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1097.512</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">E = 0.1 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">0.976</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1097.024</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">F = 0.25 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1095.56</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">G = 0.5 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">4.88</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1093.12</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">H = 1 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">9.76</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1088.24</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | </table>
| + | |
| − | <p>Mastermixes for Day 3 and Day 4: </p>
| + | |
| − | | + | |
| − | <table class="tg">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-s268">Condition</th>
| + | |
| − | <th class="tg-s268">V Pyo (uL)</th>
| + | |
| − | <th class="tg-s268">Kan (uL)</th>
| + | |
| − | <th class="tg-0lax">LB 10x uL</th>
| + | |
| − | <th class="tg-0lax">Autoclaved Water (uL)</th>
| + | |
| − | <th class="tg-0lax">Tot Vol (uL)</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-s268">A = 2.5 uM Pyo</td>
| + | |
| − | <td class="tg-s268">24.4</td>
| + | |
| − | <td class="tg-s268">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1073.6</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-s268">B = 5 uM Pyo</td>
| + | |
| − | <td class="tg-s268">48.8</td>
| + | |
| − | <td class="tg-s268">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1049.2</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">C = 10 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">97.6</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1000.4</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">D = 25 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">244</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">854</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">E = 50 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">488</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">610</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-0lax">F = 100 uM Pyo</td>
| + | |
| − | <td class="tg-0lax">976</td>
| + | |
| − | <td class="tg-0lax">2.44</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">122</td>
| + | |
| − | <td class="tg-0lax">1220</td>
| + | |
| − | </tr>
| + | |
| − | </table>
| + | |
| − | | + | |
| − | <li>Fill 96-well plate with media solutions</li>
| + | |
| − | <p>Add 100 uL of media in the desired wells, according following the label on mastermixes and table below: </p>
| + | |
| − | <table class="tg">
| + | |
| − | <tr>
| + | |
| − | <th class="tg-0pky"></th>
| + | |
| − | <th class="tg-fymr">1</th>
| + | |
| − | <th class="tg-fymr">2</th>
| + | |
| − | <th class="tg-fymr">3</th>
| + | |
| − | <th class="tg-fymr">4</th>
| + | |
| − | <th class="tg-fymr">5</th>
| + | |
| − | <th class="tg-fymr">6</th>
| + | |
| − | <th class="tg-fymr">7</th>
| + | |
| − | <th class="tg-fymr">8</th>
| + | |
| − | <th class="tg-fymr">9</th>
| + | |
| − | <th class="tg-fymr">10</th>
| + | |
| − | <th class="tg-fymr">11</th>
| + | |
| − | <th class="tg-fymr">12</th>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">A</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BA</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BA</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BA</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BA</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">B</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BB</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BB</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BB</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BB</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">C</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BC</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BC</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BC</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BC</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">D</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BD</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BD</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BD</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BD</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">E</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BE</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BE</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BE</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BE</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">F</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BF</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BF</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BF</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BF</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">G</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BG</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BG</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BG</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BG</td>
| + | |
| − | </tr>
| + | |
| − | <tr>
| + | |
| − | <td class="tg-fymr">H</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-cai3">PC1</td>
| + | |
| − | <td class="tg-0pky">BH</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-f14n">PC2</td>
| + | |
| − | <td class="tg-0pky">BH</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-xuin">-</td>
| + | |
| − | <td class="tg-0pky">BH</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-r3a5">+</td>
| + | |
| − | <td class="tg-0pky">BH</td>
| + | |
| − | </tr>
| + | |
| − | </table>
| + | |
| − | <li>OD600 matching</li>
| + | |
| − | <p>Once the cultures tubes in the shaking incubator appear cloudy, take them out and in a fresh microplate add 100 uL of each sample into a well. Add 100 uL of LB + antibiotic into a blank well. Take endpoint measurement of the filled wells and blank them with LB+antibiotic. Note down the OD600 value of the wells and dilute them to the desired initial OD600 (a value of 0.1 is suggested for a volume of 100 uL</p>
| + | |
| − | <p>Cell dilution calculator:
| + | |
| − | Volume of LB+Antibiotic to add in culture tube = [( Vcells * ODcell)/desired OD] - Vcells
| + | |
| − | E.g. to dilute 5mL of cells at an OD600 of 0.3 to OD of 0.1 = [ (5mL*0.3)/0.1 - 5] =10 mL
| + | |
| − | </p>
| + | |
| − | <li>Add cells to 96-well plate</li>
| + | |
| − | <p>Pipette 100 uL of liquid cultures ot OD600= 0.1 into the corresponding wells,following 96-well microplate layout and culture tubes label.</p>
| + | |
| − | <li>Set up the script in the plate reader and start experiment</li>
| + | |
| − | <p>For Pyocyanin experiments:</p>
| + | |
| − | <p>Set up a protocol that takes OD600 absorbance measurements and fluorescent intensity measurements every 10 minutes for 24 hours (288 cycles for 600 seconds cycles time). </p>
| + | |
| − | <p>For ferro/ferricyanide experiments:
| + | |
| − | </p>
| + | |
| − | <p>
| + | |
| − | For 200 uL of solution, we used a double orbital shaking rate of 200 rpm. Set temperature at 37°C.</p>
| + | |
| − | <li>Data analysis</li>
| + | |
| − | <p>Calculate mean and standard error for each set of replicates</p>
| + | |
| − | | + | |
| − | </ol>
| + | |
| − | </div>
| + | |
| − | <div class="clr"></div>
| + | |
| | <p2>Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat.</p2> | | <p2>Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat.</p2> |
| | </div> | | </div> |
| Line 721: |
Line 260: |
| | panelList[0].style.color = "#F76C6C"; | | panelList[0].style.color = "#F76C6C"; |
| | panelList[1].style.color = "#F76C6C"; | | panelList[1].style.color = "#F76C6C"; |
| − | panelList[1].style.transform = "rotate(180deg)" | + | panelList[1].style.transform = "rotate(0deg)" |
| | + | |
| | | | |
| | } else { | | } else { |
| Line 735: |
Line 275: |
| | } | | } |
| | | | |
| − |
| + | |
| | + | |
| | | | |
| | </script> | | </script> |