| Line 620: | Line 620: | ||
<img src="https://static.igem.org/mediawiki/2018/c/c1/T--Imperial_College--SBOL_table.png" alt="" width=“40%”> | <img src="https://static.igem.org/mediawiki/2018/c/c1/T--Imperial_College--SBOL_table.png" alt="" width=“40%”> | ||
| − | |||
</div> | </div> | ||
| Line 646: | Line 645: | ||
<p><b>Figure 3.</b>Library Assembling</p> | <p><b>Figure 3.</b>Library Assembling</p> | ||
</div> | </div> | ||
| − | |||
| Line 654: | Line 652: | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
<div type="button" class="accordion" >Summary</div> | <div type="button" class="accordion" >Summary</div> | ||
<div class="panel expt5p"> | <div class="panel expt5p"> | ||
Revision as of 23:15, 17 October 2018
Results

Building and Testing Pixcell Constructs
Assembling the Pixcell Constructs
The PixCell Construct consists of a repurposed version of the soxRS regulon from E. coli, consisting of SoxR and GFP being expressed from either side of the pSoxR/pSoxS bidrirectional promoter. pSoxR provides constitutive expression of SoxR. When oxidised, either directly by redox-cycling molecule or by oxidative stress, SoxR binds and activates transcription of GFP downstream of pSoxS.
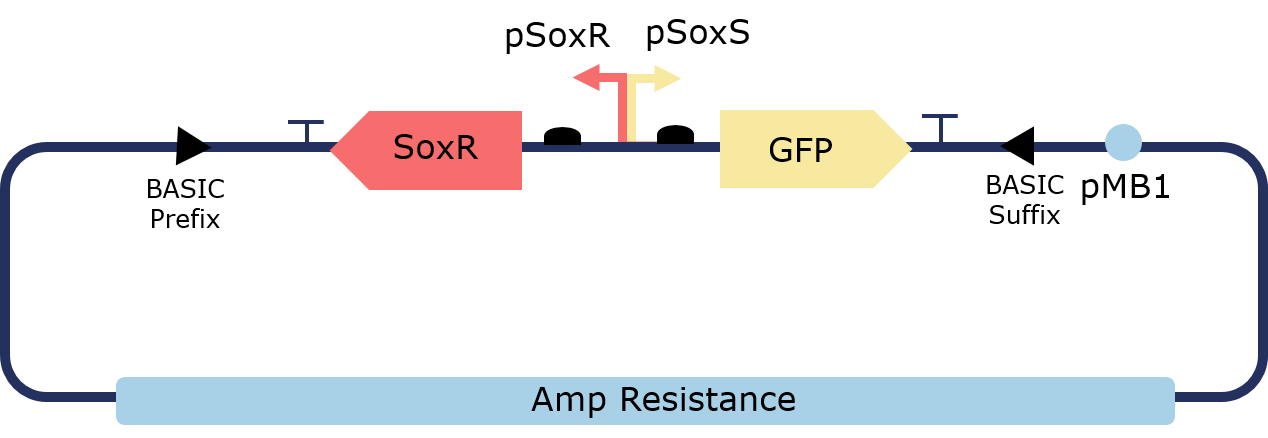
Figure 1. Illustration of the PixCell Construct
The Pixcell construct was designed to test whether we could control the expression of GFP by controlling the oxidation state of SoxR through our electrochemical set up.
Using the Golden Gate assembly method, with two inserts; the sensing portion of the soxRS regulon and GFP, which were originally amplified out from the E. coli genome and a storage vector respectively, the construct was assembled. Gel electrophoresis showed that a construct of approximately the right length was produced and the construct was then sequence verified.

Figure 1. PCR results from the amplification of the entire construct
In order to lower the basal level of GFP expression a novel degradation tag was added. Therefore, any increase in GFP expression induced by the oxidation of SoxR would be more predominant.

Figure 1. Illustration of the PixCell Construct DegTag
The PixCell construct enables us to test whether we can control gene expression by controlling the oxidation state of SoxR. The PixCell construct DegTag enables the system to be more dynamic in that seeing the system switch from on to off is quicker.
Characterizing Pixcell Constructs
We characterised the PixCell circuits in a series of steps in order to make them respond to an electronic input. In order to demonstrate if the PixCell construct is responsive to redox-cycling molecules, we started by chemically inducing the circuit with oxidised pyocyanin in solution. Characterisation experiments were performed by measuring cell growth and GFP expression at a range of increasing pyocyanin, ferricyanide and ferrocyanide concentrations, as fully described in the “Methods” section. Full growth curves and GFP expression profiles with standard error bars for each construct at each redox-molecule concentration are available in the “Supplementary Data” section.
These results demonstrate that the assembled PixCell circuit can be switched on by oxidative signals and switched off by reductive signals. These experimental data were also handed over the dry lab, where information about fold induction and degradation rates supported and further implemented the theoretical models.
As a first step, we investigated the effect of oxidised pyocyanin on cell growth. We observed that pyocyanin concentrations higher than 10 uM significantly impacted cell growth (figure 1). We then measured GFP expression at a range of different concentrations (figure 2). We found an optimal trade-off between cell growth and GFP expression at a concentration of 2.5 uM, which is half of that used by (Tschirhart et al., 2017) in their anaerobic system. Once identified our working pyocyanin concentration we varied the concentration of ferro/ferricyanide, the modulators of cellular redox activity. As above, we investigated the effect of the redox molecules on cells containg the PixCell Constructs in order to identified a ferro/ferricyanide concentration (10 mM), which provided optimal balance between GFP expression and cell growth.




Figure 1.Pyocyanin affects cell viability




Figure 2.Pyocyanin induces GFP expression

Figure 3.Pyocyanin dose response

Figure 4.Inorganic sulfite plates

Figure 5.Production of pyocyanin sulfonate

Figure 6.Final sulfite concentration


Figure 7.FCN dose responses

Figure 8.Final Pyocyanin, ferrocyanide, ferricyanide and sulfite concentrations in agar plates
These experiments allowed us to demonstrate that our constructs are responsive to oxidising input and enabled us to identify the concentrations of redox-molecule. This allowed us to select a final chemical condition of 2.5uM Pyo, 0.02% sodium sulfite and 10mM ferrocyanide.
Electrochemistry of Redox Modulators
To demonstrate the ability of our electrode set-up to selectively oxidise and reduce our redox modulators in solution, we investigated the redox status of the our redox molecules with square wave voltammetry (SWV) and by measuring the absorbance at 390 nm with a plate reader. SWV data showed that a voltage of +0.5 at the working electrode enables bulk oxidation, whereas a voltage of -0.4 allows bulk reduction. However, the presence of oxygen in aerobic environments interferes with the oxidation signal carried by the electrodes. Measuing optical absorbance at 390 nm with a plate reader, we then investigated the redox kinetics of pyocyanin in the presence of the reducing agent sodium sulfite.
These results show that our set-up is able to selectively and reversibly oxidise and reduce our redox modulators. Furthermore , by performing these electrochemistry experiments, we proved that addition of the reducing agent sodium sulfite enables complete chemical reduction of the system, even in aerobic environment.

Figure 1. Square-wave voltammetry setup
The above setup was used to perform Square-wave voltammetry to investigate bulk oxidation and reduction of our system. The setup was controlled using a multichannel potentiostat connected to a computer.

Figure 2. Square-wave voltammetry of redox modulator
The graphs above summarise the results showing that our system was consistently bulk reduced at -0.3V and bulk oxidised at +0.5V. The addition of Sulfite and cell containing the pixCell construct did not significantly alter the reduction and oxidation profile. These results led us to chose +0.5V as our oxidising potential in the following experiments testing the spatial electronic control of gene induction.

Figure 3. Amperometry of the final plate conditions
Amperometry was performed on plates containing 2.5uM Pyocyanin, 10mM FCN(R), Ampicillin, Kanamycin, 0.02% sulfite and E. coli containing PixCell construct Deg Tag. The results show that the current decays exponentially until a steady state is reached indicating bulk oxidation of redox modulators.
These results describe the first ever aerobic electrogenetic control system, that works both in liquid and solid E. coli cultures.
Spatial Electronic Control of Gene Induction
Here we demonstrated the first spatial activation of gene expression using electronic control.
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
An electrode rig was set up to apply these potentials to cells grown on an agar plate containing the final reaction condition of 2.5μM pyocyanin, 0.02% sodium sulfite and 10mM ferrocyanide. Redox reactions only occur at the electrode surface during an electrochemistry experiment, meaning that oxidised pyocyanin and ferrocyanide were only produced in close proximity to the working electrode upon the application of a +0.5V pulse. Fluorescence images of agar plates clearly show localised expression of GFP around the electrode. This not only shows that the device allows for electronic control of gene expression, but also demonstrates high spatial control meaning it can be used for programmable spatial patterning of cell populations.

Figure 1. Setup of the electrode rig for the electronic control of spatial gene induction

Figure 2. Image of the electrode rig placed in the incubator.
Figures 1 and Image 2 illustrate the setup of the electrode rig. The working, counter and reference electrodes are penetrating the plate through holes. Cells were plated onto an agar plate containing the final working concentrations: 10mM FCN(R), 2.5 uM pyocyanin, 0.03% sulfite, ampicillin and kanamycin. Agar is fully covering the electrodes which are controlled by a potentiostat. An oxidising potential of +0.5% was applied.

Figure 3. Expected outcome for the electronic control of gene expression on an agar plate
Figure 3 illustrates the expected increased fluorescence around the working electrode. The oxidising potential results in bulk oxidation and activation of our system.
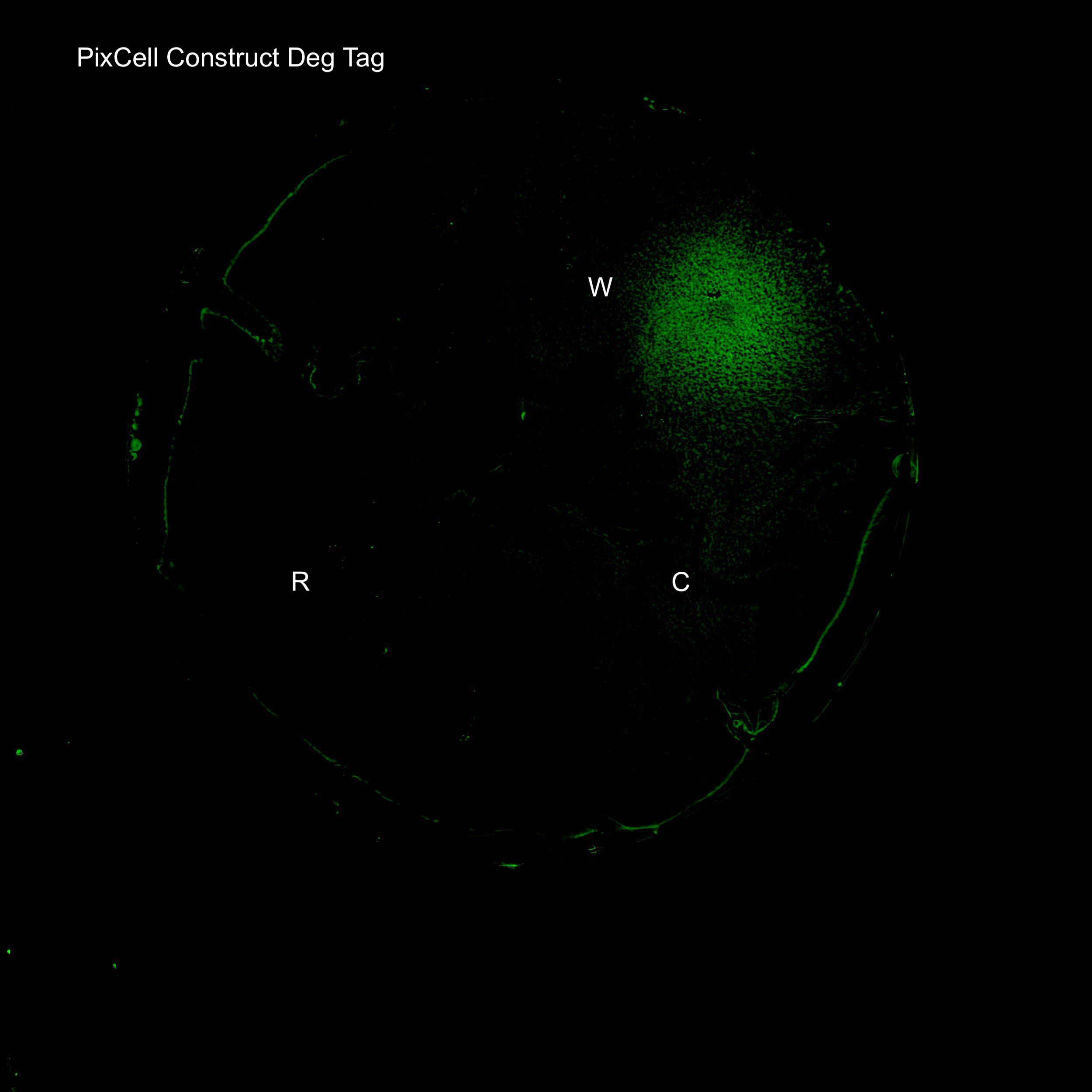
Figure 4. GFP expression after electric stimulation
We demonstrated elevated GFP expression around the working electrode in figure 4.

Figure 5. Comparison of PixCell Construct, PixCell Construct Deg Tag, positive control and negative control after application of an oxidising potential
We next investigated GFP expression of PixCell Construct and PixCell Construct Deg Tag relative to the negative control (DJ901) and the positive control which constitutively expresses GFP. Fluorescence image analysis shows increased fluorescence around the working electrodes demonstrating electronic control of spatial gene expression. The addition of a degradation tag to GFP substantially reduces background fluorescence.
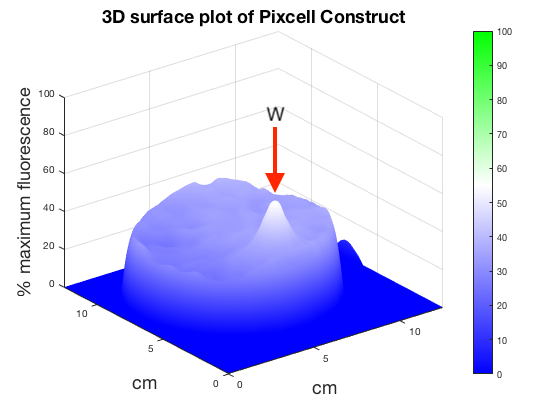



Figure 6. 3D surface plot showing relative fluorescence of PixCell Construct, PixCell Construct Deg Tag, positive control and negative control after electric stimulation
To better visualise the results 3D surface plots were made showing GFP expression in the z axis. GFP expression level from the positive and negative control was used to set a relative scale of GFP expression. Peaks around the working electrodes clear indicate increased GFP expression. PixCell construct Deg Tag showed lower background fluorescence compared to PixCell construct.

Figure 7. Comparison of PixCell Construct, PixCell Construct Deg Tag, positive control and negative control after application of a reducing potential
Next the 4 cell types were exposed to a reducing potential of -0.3V. Working electrodes do not show increased fluorescence. Higher background fluorescence in the plate with cells containing PixCell Construct compared to the the previous fluorescence images exposed to an oxidising potential is due to variation in spreading.

Figure 8. GFP expression on a plate with final conditions in the absence of pyocyanin
After successfully inducing spatially controlled activation of gene expression the system was tested in the absence of the redox modulator pyocyanin. The plate contains 10mM FCN(R), 0.03% sulfite, Ampicillin, Kanamycin. Results show that increased expression levels of GFP can be achieved around the working electrode in the absence of pyocyanin. Further fine tuning of the system will be required to achieve the level of precision obtained on a plate containing our final working conditions.
These results demonstrate precise electronic control of spatial gene expression, in solid cultures grown in aerobic condition.
Building and Characterizing the Pixcell Library
Creating the Sox Library
The PixCell part library consists of 5 SoxR transcription factors and 8 pSoxS promoters which were created for construction of electrogenetic circuits with variable activity. This allows for control of this system on both the genetic and the electrochemical layer. SoxR transcription factors were obtained from a series of homologues from non-pathogenic species. pSoxS was completely rengineered as a promoter. The promoter was made unidirectional by knocking out the upstream constitutively active pSoxS portion, a terminator was placed upstream to allow for efficient tandem assemblies and a downstream ribozyme was added to remove context dependency. Promoters were either sourced from homologues in non-pathogenic bacteria or were produced by conservative mutations to the consensus sequences and SoxR binding site. The library includes a mutant promoter which provides transcriptional repression rather than activation.
These results expand the electrogenetic synthetic biology toolkit.

Figure 1.Library design

Figure 2.OD600 of parts over time

Figure 3.Library Assembling
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Characterising the Sox Library
All assembled constructs were transformed into DH5-alpha with single colonies being grown for 15 hours with cells being grown in LB with either 0μM and 2.5μM pyocyanin. The normalised fluorescence from each cell was then calculated and compared. Firstly all fluorescence values were divided by the fluorescence of the library seed (BBa_K##### + BBa_K######) which is made from our versions of the native E. coli SoxR and pSoxS. This gave an RPU scale giving a range of promoter activities between X-X RPUs, exhibiting tunability of our system.

Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Figure 1. Pyocyanin affects growth in cells containing the library seed construct

explain
Figure 1. Pyocyanin affects growth in cells containing the library seed construct



explain
Figure 2.Pyocyanin induces GFP expressionin cells containing the library seed construct



Taking the steady state fluorescence values we can show GFP expression (and therefore construct activity) as a function of pyocyanin activity. This shows a standard sigmoidal behaviour as predicted by our model and exhibited by PixCell Patterning Construct 1 (BBa_######).
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Testing Alternative Non-toxic Redox Modulators
Testing Phenazine Methosulfate
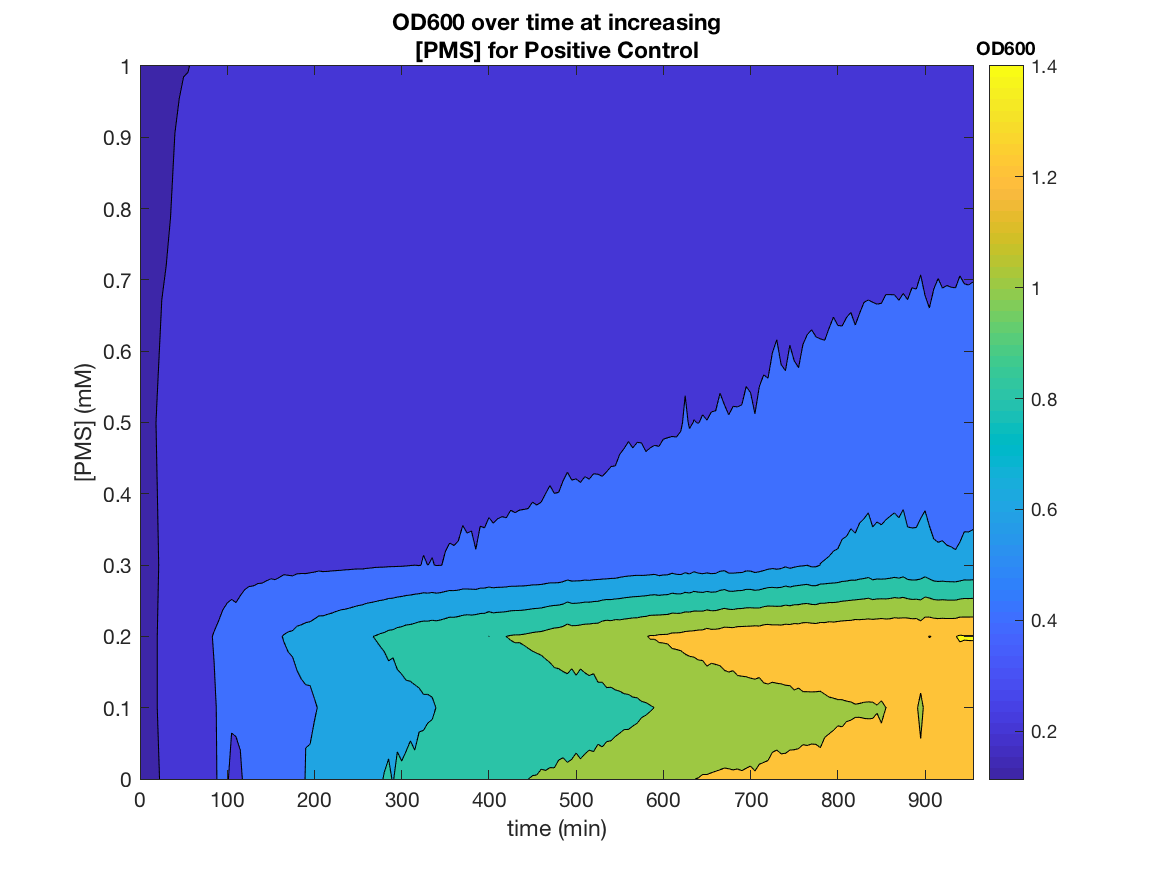
Our electrogenetic control system relies on the electron carrying action of the redox-cycling drug pyocyanin, which is toxic metabolite from synthesised by Pseudomonas aeruginosa. This poses limitations to our system, with regards to safety and possible applications. Therefore, we searched in the literature for alternative SoxR inducers and and tested potential non-toxic alternatives. Shown below are the characterization data for the best candidate alternative: phenazine, methosulfate.
These results demonstrate the first ever use of PMS as an electrogenetic inducer.
The above setup was used to perform Sqaure-wave voltammetry confirming that ferrocyanide was bulk reduced at -0.3V and bulk oxidised at +0.5V.
Figure 1. Phenazine as as Cheaper, Non-toxic Redox Modulator

Figure 2. Phenazine effects on cell growth


explain
Figure 5. Phenazine induces GFP expression in PixCell Patterning Circuits



explain
These results demonstrates the SoxR/pSoxS system that we devised, can be used a non-toxic and effective tool for chemical, as well as electronic, gene induction. PMS is also incredibly cheap, being 40,000x cheaper than pyocyanin, ~190x cheaper than arabinose and ~29x cheaper than aTc per reaction.
Applications
Biocontainment- Gp2 Growth Inhibition
In order to test our PixCell electrogenetic part library we constructed a biocontainment device to prototype one of our proposed applications. This was constructed using an existing part in the iGEM registry: the growth inhibitor Gp2 (BBa_K1893019). By placing this downstream of pSoxS (BBa_K2862006) and expressing SoxR (BBa_K2862014) from a constitutive promoter (BBa_K2862019) we could inhibit cell growth in the oxidising conditions we produced from our electrochemical set up. We envisioned this being used with our array in a biocontainment device in which any cells that spread to the edges of the device have their growth seized by an oxidising potential. This device could be used to prevent the accidental release of GMOs, much like how electric fences are used to restrain animals.
These results serve as proof of concepts for the biocontainment application, as described in the “Applications” sections
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat. Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Figure 1.Gp2 test growth curves
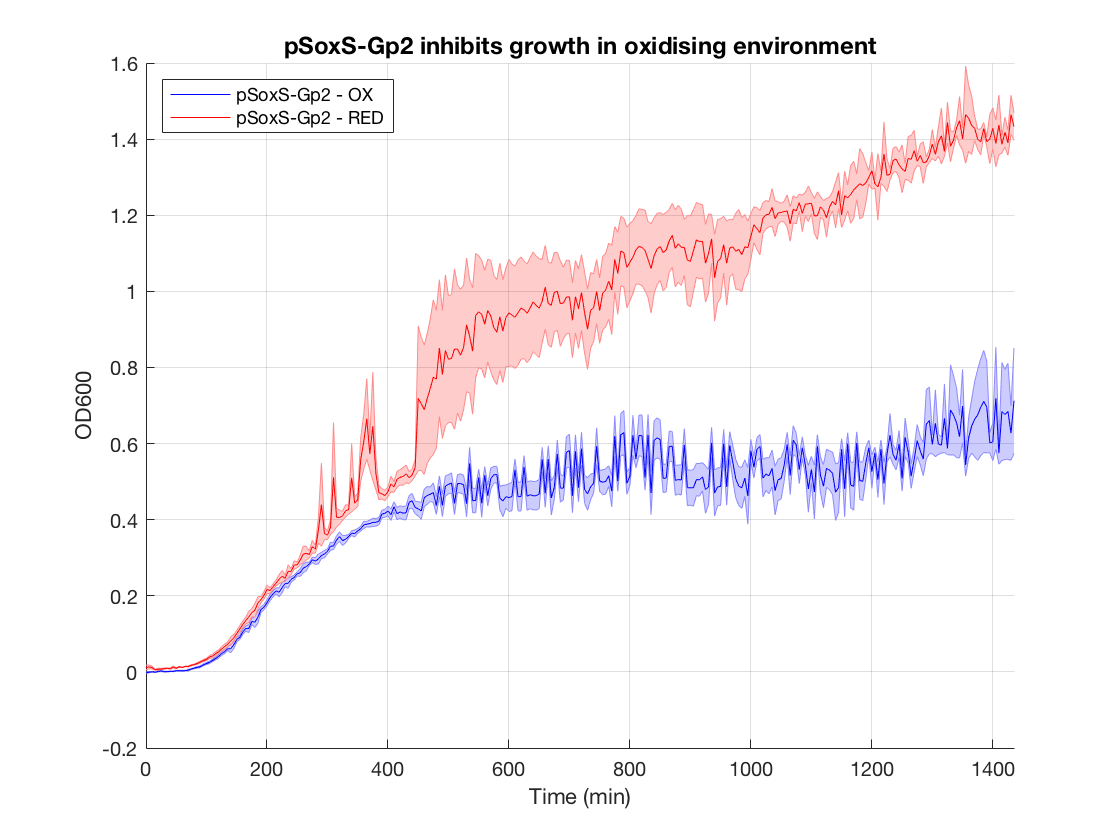
Figure 2.Gp2 control growth curves

Figure 3.Gp2 bar chart

When looking at the steady state OD600 values of these constructs a clear significant ~2-fold reduction is seen in the application construct in oxidising conditions. Furthermore there is no difference between the growth of the reducing condition and the controls. This not only provides a proof-of concept for out application, but also proves the system is not significantly leaky. This means it is feasible to use a higher copy number plasmid or more toxic gene (such as MazF) in this device to provide an even more effective biocontainment device. .
With some improvement, and when used in combination with the PixCell electrode array, this biocontainment device could be used for biocontainment of GMOs in contained-use devices.
Fabric Bioprinting - Melanin
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat. Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.

Figure 1. PCR results of genomic extraction of SoxR from E. coli (including bi-directional promoter and SoxR protein gene)
Lorem ipsum dolor sit, amet consectetur adipisicing elit. Excepturi, nisi illum. Consequuntur cum, sequi quo, esse facere corrupti voluptatum ipsum vel consequatur error quasi tenetur, repellat voluptatibus aspernatur optio mollitia! Lorem ipsum dolor sit, amet consectetur adipisicing elit. Excepturi, nisi illum. Consequuntur cum, sequi quo, esse facere corrupti voluptatum ipsum vel consequatur error quasi tenetur, repellat voluptatibus aspernatur optio mollitia!
Lorem ipsum dolor, sit amet consectetur adipisicing elit. Recusandae dolore, tenetur expedita in architecto, repellat rerum minima cumque exercitationem debitis laborum fugit aliquam quo molestiae. Alias eius nostrum aspernatur quaerat.













