heading
Our Results
Identification of IL7-AS expression in renal cell carcinoma by TANRIC
Based on evidence that IL7-AS can be induced across multiple human and mouse cell types and has been reported to regulate the expression of interleukin-6 (IL6). IL6 is emerging as a major regulator of renal cell cancer. We asked whether IL7-AS is involved in pathogenesis of renal cell carcinoma. By analyzing data from TANRIC (http://ibl.mdanderson.org/tanric/_design/basic/query.html), we collected the expression data of IL7-AS in 64 normal tissues and 432 kidney renal clear cell carcinoma tissues. IL7-AS was expressed at higher levels in kidney renal clear cell carcinoma (KIRC) than in normal kidney (Fig. 1).

Fig. 1 IL7-AS was expressed at higher levels in kidney renal clear cell carcinoma than in normal kidney by TANRIC.
Identification of IL7-AS expression in kidney cancer cell lines
The 769-P and 786-O cell lines are derived from primary clear cell adenocarcinoma. The 293T cell line is originally derived from human embryonic kidney. We examined the expression levels of IL7-AS in 769-P, 786-O and HEK-293T cell lines. IL7-AS expression was markedly higher in 786-O and 769-P cells than in HEK-293T cells (Fig. 2). We also observed that up-regulated IL7-AS expression was correlated with IL6 expressions in these cell lines (Fig. 2).
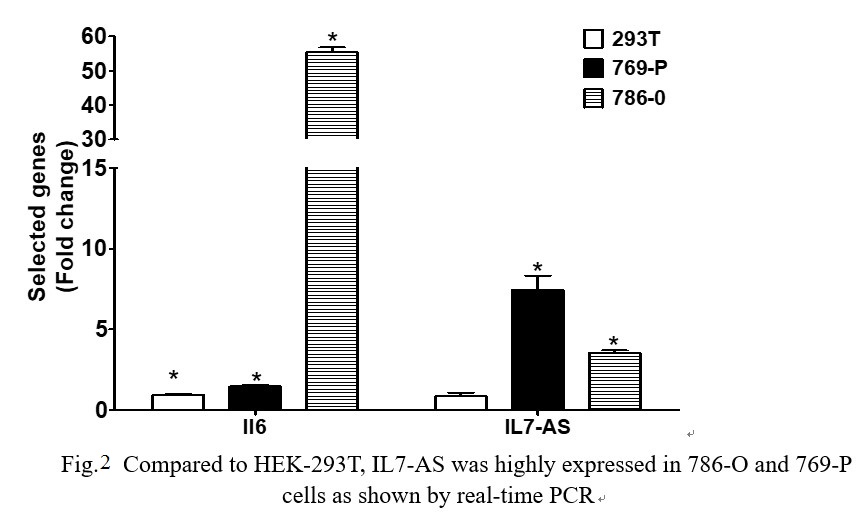
Fig. 2 Compared to HEK-293T, IL7-AS was highly expressed in 786-O and 769-P cells as shown by real-time PCR
IL7-AS promotes kidney cancer cell migration
Cell migration is a central process in the development and maintenance of tumor. We cloned the full length of IL7-AS, two truncated sequences of IL7-AS (IL7-AS-S1 and IL7-AS-S2) into PCDNA3.1 (Fig. 3) and transfected these plasmids to 786-O cells. Through in vitro scratch wound healing assay, overexpression of IL7-AS promoted cell migration of 786-O cells (Fig. 4 and 5). In order to determine which domain is essential for the migration of IL7-AS, we cloned two truncated sequences of IL7-AS (IL7-AS-S1 and IL7-AS-S2). Overexpression of IL7-AS-S1 or IL7-AS-S2 increased the migration of 786-O cells (Fig. 4 and 5). These results suggested that IL7-AS-S1 and IL7-AS-S2 contain key structural domains. IL7-AS may play an important role in migration of renal cell carcinoma.
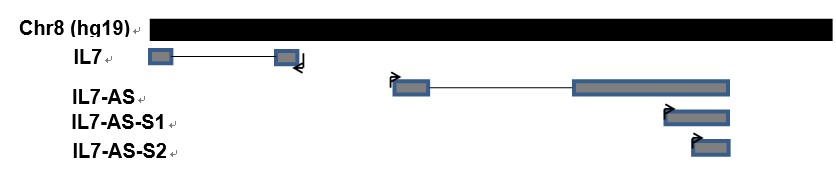
Fig. 3 Scheme of three IL7-AS isoforms on human chromosome 8
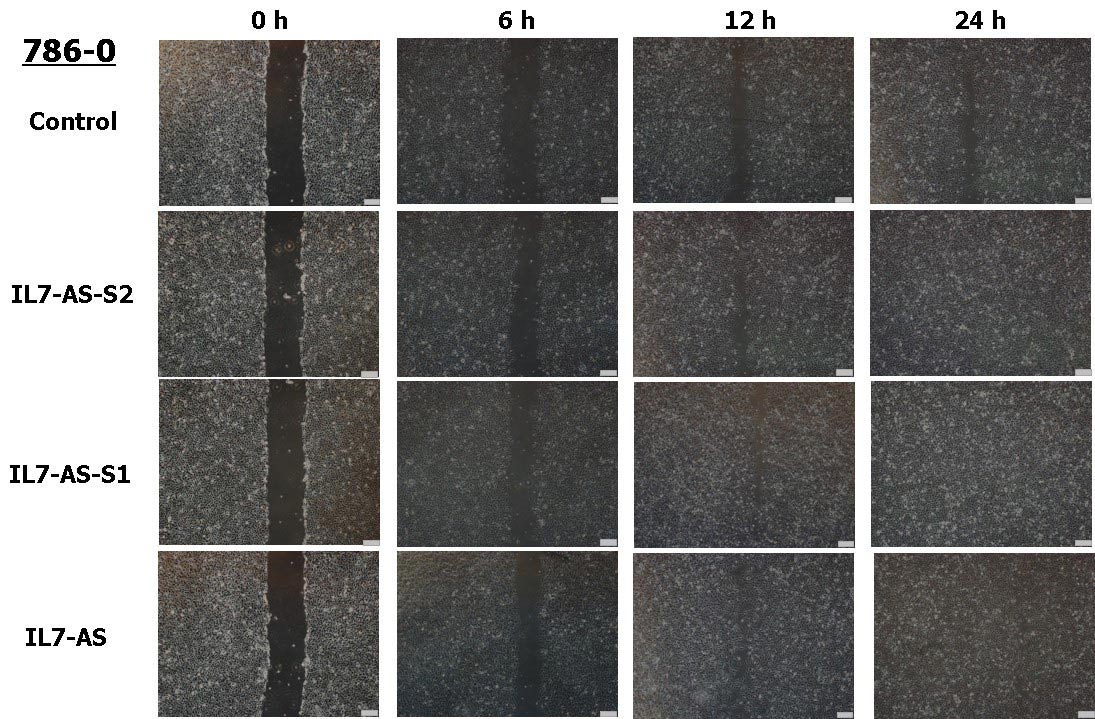
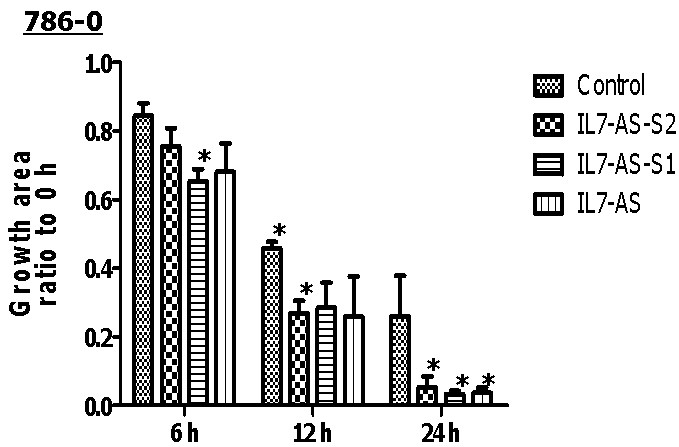
Fig. 4 Overexpression of IL7-AS-S2, IL7-AS-S1 and IL7-AS promotes 786-O cells migration. 786-O cells were transfected with pcDNA3.1-IL7-AS-S2, pcDNA3.1-IL7-AS-S1 or pcDNA3.1-IL7-AS for 24 h. The cells growth areas were calculated with Image J at the time points of 0, 6, 12 and 24 h. Bar=100 μm. Data represents means ± SE from 3 independent experiments. *p<0.05 versus non-transfected cells.
In order to investigate whether cell migration induced by IL7-AS-S2 depends on the amount of IL7-AS-S2 transfection. We transfected the different concentrations of IL7-AS-S2 (1ug, 2ug, 4ug) into 786-0 cells. Both three concentrations of IL7-AS-S2 can infect the migration of 786-0 cells (Fig. 5).
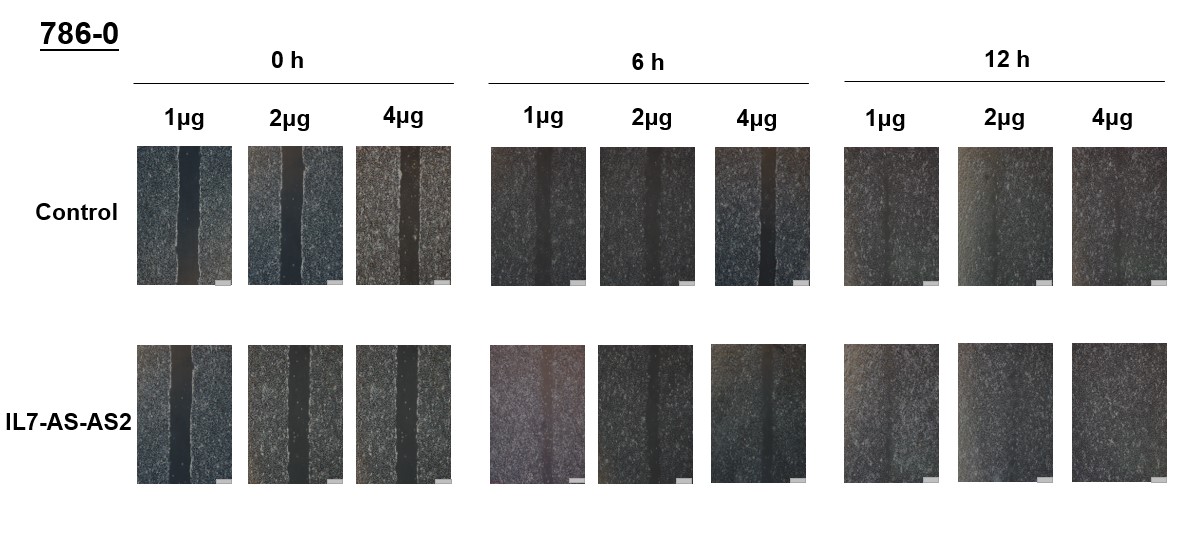
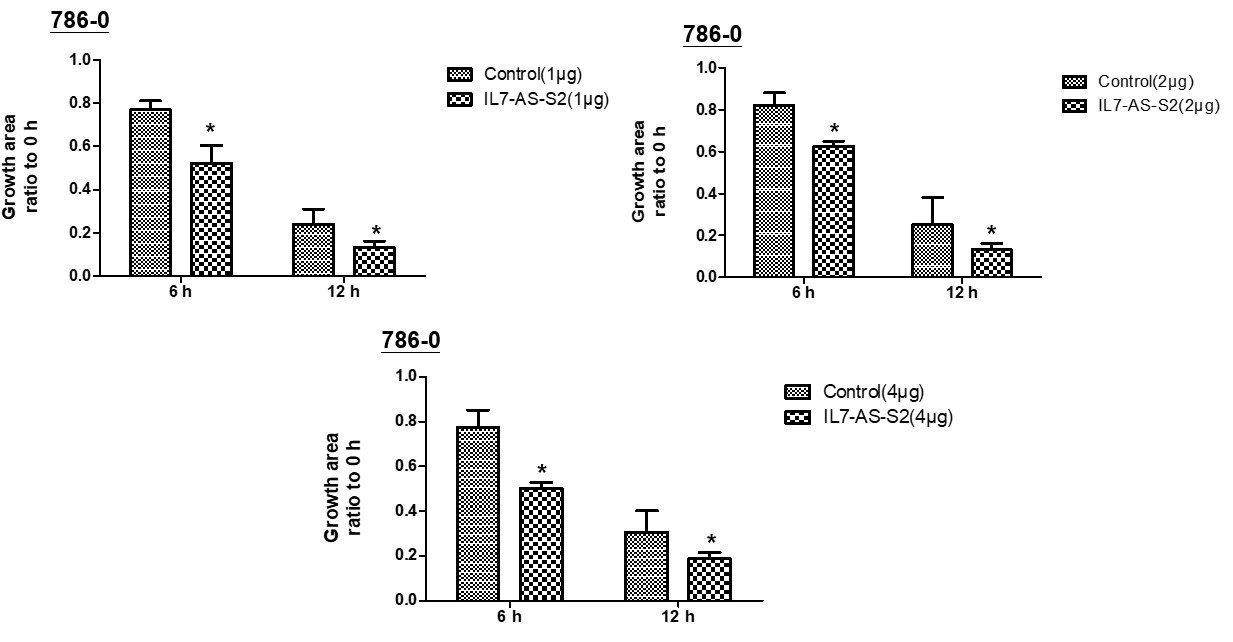
Fig. 5 Different concentrations of IL7-AS-S2 promotes 786-O cells migration. 786-O cells were transfected with pcDNA3.1-IL7-AS-S2 (1 ug, 2 ug, 4 ug) for 24 h. The cells growth areas were calculated with Image J at the time points of 0, 6, and 12 h. Bar=100 μm. Data represents means ± SE from 3 independent experiments. *p<0.05 versus non-transfected cells.
IL7-AS contains a complicated secondary structure
In silico prediction of lncRNA secondary structure is another useful method to assign putative functions to non-coding transcripts, based upon the widely held assumption that highly folded structures impart functionality through binding interactions with proteins/nucleotides. Characterization of IL7-AS, IL7-AS-S1 and IL7-AS-S2 using RNA fold minimum free energy estimations predicted a highly folded secondary structure with several hairpin loops, which imply interact with protein (Fig. 6).
A. IL7-AS
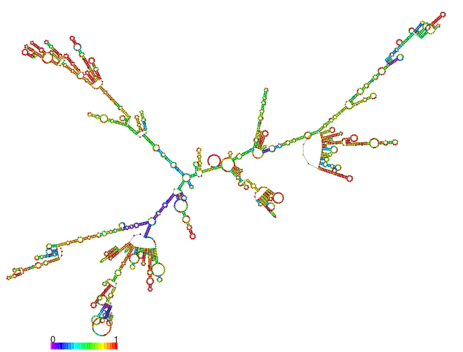
B. IL7-AS-S1
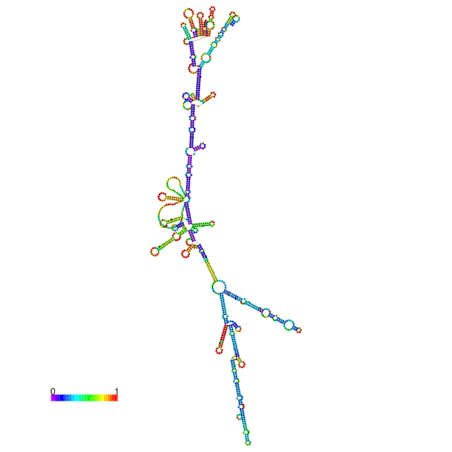
C. IL7-AS-S2
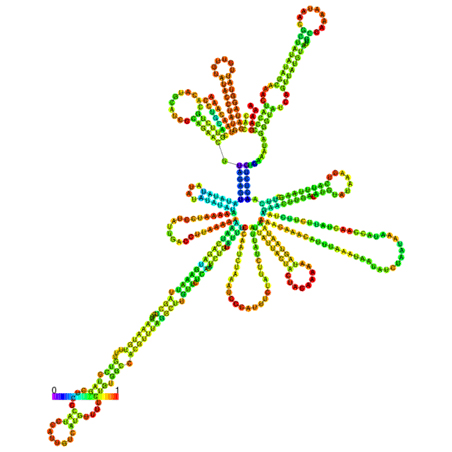
Fig. 6 The optimal secondary structure with a minimum free energy. The secondary structures of IL7-AS; IL7-AS-S1 and IL7-AS-S2 were predicted by the RNAfold webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The structure is colored according to base-pairing probabilities. For unpaired regions, the color denotes the probability of being unpaired.
Bioinformatic prediction of IL7-AS-S2 function
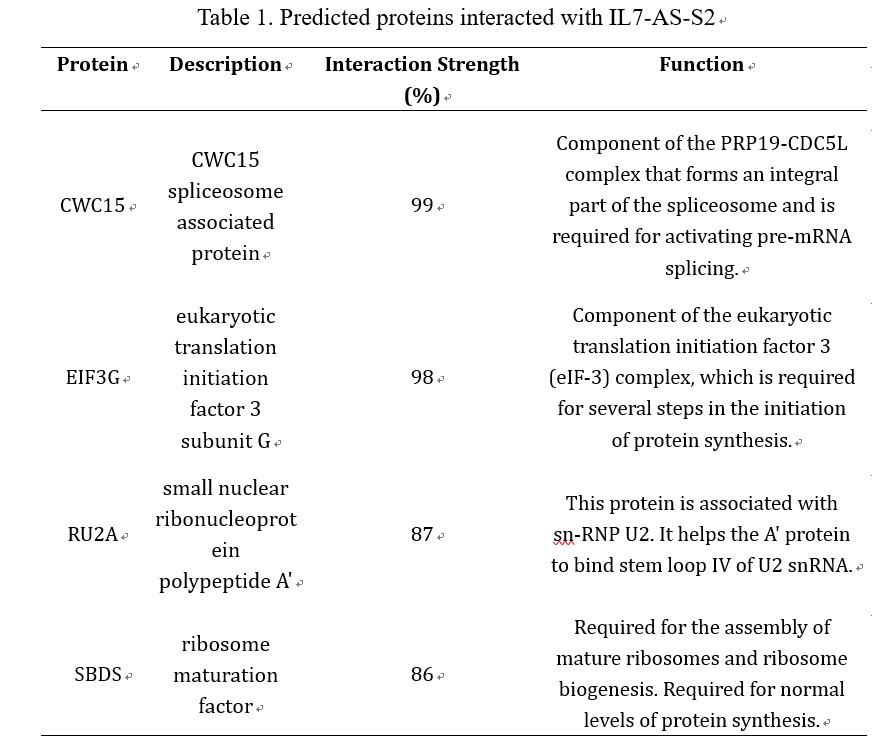
LncRNAs perform the function through interacting with proteins. Considering IL7-AS-S2 can influence the cell migration, we want to predict proteins which can interact with IL7-AS-S2. Using the catRAPID omics algorithm (http://service.tartaglialab.com/page/catrapid_omics_group), we determined whether IL7-AS-S2 interacts with proteins. Results showed that the related RNA-binding proteins were involved in RNA-splicing/processing (CWC15, RU2A), and protein synthesis (EIF3G, SBDS) (table 1). Based on the proteins predicted, we estimated that IL7-AS-S2 may regulate RNA splicing or protein synthesis of some tumor related genes though interacting with several RNA-splicing or protein synthesis associated proteins.
Table 1. Predicted proteins interacted with IL7-AS-S2
Conclusions and perspectives
The results reported here demonstrate that IL7-AS may play an important role in pathogenesis of renal cell carcinoma. We found that IL7-AS was expressed at higher levels in kidney renal clear cell carcinoma than in normal kidney. Moreover, IL7-AS expression was higher in renal cell carcinoma associated cells than in human embryonic kidney cells. IL7-AS promoted the migration of renal cell carcinoma related cells. Our results for the first time to evaluate the possibility of IL7-AS in renal cell carcinoma as a novel biomarker for diagnosis and a potential target for therapy in the future.

