| Line 55: | Line 55: | ||
<h2>Eric's Latex files (to insert)</h2> | <h2>Eric's Latex files (to insert)</h2> | ||
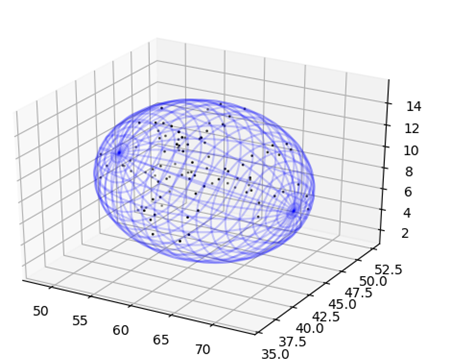
<p>We determined that the linkers between the GR-LBD and the inteins halves would need to be flexible as | <p>We determined that the linkers between the GR-LBD and the inteins halves would need to be flexible as | ||
| − | + | there is no direct path connecting the termini and they would require the flexibility to meet each other | |
| − | + | and produces a trans-splicing event. To determine which linker would be ideal we ran simulations of | |
| − | + | Root-mean-square deviation of atomic positions to determine optimal linker length and composition. | |
| − | + | Firstly, we chose various linker designs inspired from previous work [a,b], then the entropy equations | |
| − | + | were run on the Queen’s University Computer Cluster for seven days by Dr. Campbell. | |
| − | < | + | <ol type="a"> |
| − | <li></li> | + | <li><a href="http://parts.igem.org/Protein_domains/Linker">http://parts.igem.org/Protein_domains/Linker</a></li> |
| − | <li></li> | + | <li><a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3726540/">https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3726540/</a></li> |
| − | </ | + | </ol> |
</p> | </p> | ||
| − | + | <p>Another aspect of our project we sought to model was which pacifier design would allow for optimal detection of the luminescence signal. | |
| − | + | Eric modelled this through a MATLAB simulation of the “Narrow escape problem”.... | |
| + | </p> | ||
| + | <p>Lastly, the analysis was modeled by… | ||
| + | </p> | ||
| + | |||
| + | <h5>Summary</h5> | ||
| + | <p>As a part of our construct it is necessary to build linkers to connect the intein halves with the target receptor. The challenge in | ||
| + | developing linkers for the system is that they must be of a specific length that will allow association of the intein halves in the | ||
| + | bound conformation of the receptor but will not allow association of the intein halves in the unbound conformation of the receptor. | ||
| + | In addition, the flexibility of the linkers must be adjusted for the same purpose. In comparison, the scientific paper from which our | ||
| + | project draws its inspiration has a much simpler time developing linkers as the change in confirmation of their chosen receptor created | ||
| + | a significantly larger change in distance compared to the receptor that we are looking at. As the project progressed many different | ||
| + | attempts at creating linkers were made. | ||
| + | </p> | ||
| + | |||
| + | <h5>Approaches - Initial Design</h5> | ||
| + | <p>The initial linkers that we wanted to test were from the scientific paper that was used for our project inspiration. As there is a high | ||
| + | degree of homology between the receptor used in the paper and the receptor that we were interested in we decided to try the exact linkers | ||
| + | used in the paper. This approach was flawed however as it overlooked the differences in the receptor conformation changes. | ||
| + | </p> | ||
| + | <!-- image with caption --> | ||
| + | |||
| + | <h5>Approaches - Uniformed Design</h5> | ||
| + | <p>While testing the initial design it was also decided to test other linkers at the same time. These linkers were generated by using common | ||
| + | synthetic linkers, found through research, and testing to see if an association event was viable by modelling the system on PyMOL. This | ||
| + | approach generated linkers that were too long as we were focussed on making linkers long enough for the interaction but forgot about the | ||
| + | need for the linkers to prevent interaction in the unbound conformation. | ||
| + | </p> | ||
| + | <!-- image with caption --> | ||
| + | |||
| + | <h5>Approaches - Informed Design</h5> | ||
| + | <p>New linkers were developed by discussing with Professors at Queen’s University for advice on linker design. As such were made aware of more | ||
| + | variables to consider when designing the linker including flexibility and secondary structure. | ||
| + | </p> | ||
| + | <!-- image with caption --> | ||
| + | |||
| + | <h5>Approaches - Modelled Design</h5> | ||
| + | <p>To try to make more informed linkers we talked with another Professor at Queen’s University. From this discussion we discussed the shortest | ||
| + | path around the molecule that would be ideal for the association event. We were also provided with web databases that contain the structure and | ||
| + | information for synthetic and natural linkers. In PyMOL the angstrom distances on the newly chosen path were determined and linkers with the | ||
| + | appropriate length and flexibility were made. | ||
| + | </p> | ||
| + | <!-- image with caption --> | ||
| + | |||
| + | <h5>Approaches - Software Design</h5> | ||
| + | <p>To further the work done to design linkers a software program that interacts with PyMOL is being developed to find the shortest path for the | ||
| + | user and design a linker that will span the space. | ||
| + | </p> | ||
| + | |||
</body> | </body> | ||
</html> | </html> | ||
Revision as of 02:34, 2 September 2018
Linker Dynamics (Abigael)
The software being developed is designed to connect two selected points on the molecule by finding the shortest path that connects them without interfering with the rest of the molecule. This is done by first finding the shortest path at a specified resolution using an extended version of Dijkstra’s Algorithm. After the shortest path has been found linkers are designed to suit the path found. The method used to generate the linkers has yet to been determined. This software will be usable through the PyMOL software to account for visual representation and interaction with the molecule of choice.


Eric's Latex files (to insert)
We determined that the linkers between the GR-LBD and the inteins halves would need to be flexible as there is no direct path connecting the termini and they would require the flexibility to meet each other and produces a trans-splicing event. To determine which linker would be ideal we ran simulations of Root-mean-square deviation of atomic positions to determine optimal linker length and composition. Firstly, we chose various linker designs inspired from previous work [a,b], then the entropy equations were run on the Queen’s University Computer Cluster for seven days by Dr. Campbell.
Another aspect of our project we sought to model was which pacifier design would allow for optimal detection of the luminescence signal. Eric modelled this through a MATLAB simulation of the “Narrow escape problem”....
Lastly, the analysis was modeled by…
Summary
As a part of our construct it is necessary to build linkers to connect the intein halves with the target receptor. The challenge in developing linkers for the system is that they must be of a specific length that will allow association of the intein halves in the bound conformation of the receptor but will not allow association of the intein halves in the unbound conformation of the receptor. In addition, the flexibility of the linkers must be adjusted for the same purpose. In comparison, the scientific paper from which our project draws its inspiration has a much simpler time developing linkers as the change in confirmation of their chosen receptor created a significantly larger change in distance compared to the receptor that we are looking at. As the project progressed many different attempts at creating linkers were made.
Approaches - Initial Design
The initial linkers that we wanted to test were from the scientific paper that was used for our project inspiration. As there is a high degree of homology between the receptor used in the paper and the receptor that we were interested in we decided to try the exact linkers used in the paper. This approach was flawed however as it overlooked the differences in the receptor conformation changes.
Approaches - Uniformed Design
While testing the initial design it was also decided to test other linkers at the same time. These linkers were generated by using common synthetic linkers, found through research, and testing to see if an association event was viable by modelling the system on PyMOL. This approach generated linkers that were too long as we were focussed on making linkers long enough for the interaction but forgot about the need for the linkers to prevent interaction in the unbound conformation.
Approaches - Informed Design
New linkers were developed by discussing with Professors at Queen’s University for advice on linker design. As such were made aware of more variables to consider when designing the linker including flexibility and secondary structure.
Approaches - Modelled Design
To try to make more informed linkers we talked with another Professor at Queen’s University. From this discussion we discussed the shortest path around the molecule that would be ideal for the association event. We were also provided with web databases that contain the structure and information for synthetic and natural linkers. In PyMOL the angstrom distances on the newly chosen path were determined and linkers with the appropriate length and flexibility were made.
Approaches - Software Design
To further the work done to design linkers a software program that interacts with PyMOL is being developed to find the shortest path for the user and design a linker that will span the space.
