JimmyChung (Talk | contribs) |
JimmyChung (Talk | contribs) |
||
| Line 6: | Line 6: | ||
<style> | <style> | ||
body { | body { | ||
| − | + | background-color: white; | |
| − | + | font-family: Helvetica, sans-serif; | |
| − | + | margin-top: 20px; | |
| − | + | position: relative; | |
| − | + | margin-left: auto; | |
| − | + | margin-right: auto; | |
} | } | ||
figure { | figure { | ||
| − | + | font-style: italic; | |
| − | + | font-size: smaller; | |
| − | + | text-indent: 0; | |
| − | + | margin-top: 10px; | |
} | } | ||
figcaption { | figcaption { | ||
| − | + | margin-top: 10px; | |
} | } | ||
</style> | </style> | ||
| Line 26: | Line 26: | ||
<body> | <body> | ||
| − | <div style="width: | + | <div style="width:70%;margin-left:15%"> |
| − | + | <h2>Linker Software Summary</h2> | |
| − | + | <h5>Summary</h5> | |
| − | + | <p>The software being developed is designed to connect two selected points on the molecule by finding the | |
| − | + | shortest path that connects them without interfering with the rest of the molecule. This is done by first | |
| − | + | finding the shortest path at a specified resolution using an extended version of Dijkstra’s Algorithm. | |
| − | + | After the shortest path has been found linkers are designed to suit the path found. The method used to | |
| − | + | generate the linkers has yet to been determined. This software will be usable through the PyMOL software | |
| − | + | to account for visual representation and interaction with the molecule of choice.</p> | |
| − | + | ||
| − | + | <h5>File Description</h5> | |
| − | + | <h6>Main File</h6> | |
| − | + | <p>The main file interfaces with PyMOL allowing for user inputs to be used in the linker software. | |
| − | + | In addition, this file processes the inputs and manages all the function calls that coordinate the creation of the linkers.</p> | |
| − | + | <figure> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/6/67/T--Queens_Canada--PyMOL.jpg" alt='pymol'/> | |
| − | + | <figcaption>An image of the PyMOL model of the molecule used for initial testing and demonstration.</figcaption> | |
| − | + | </figure> | |
| − | + | ||
| − | + | <h6>Ellipsoid Fit</h6> | |
| − | + | <p>The ellipsoid fit module finds the tightest fitting ellipsoid to a set of points that represent the | |
| − | + | atoms that compose the molecule. This is done using an algorithm that is based on the Khachiyan Algorithm. This module returns the | |
| − | + | center, radii and rotation of the ellipsoid that encloses the points and allows for the plotting of the ellipsoid as a mesh graph.</p> | |
| − | + | <figure> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/7/72/T--Queens_Canada--BestFitEllipsoid.jpg" alt='bestfit'/> | |
| − | + | <figcaption>The best fitting ellipsoid to the set of points. The set of points that represent the atoms comprising | |
| − | + | the molecule are shown above as black dots while the best fitting ellipsoid is represented as a blue mesh grid.</figcaption> | |
| − | + | </figure> | |
| − | + | ||
| − | + | <h6>Ellipsoid Points</h6> | |
| − | + | <p>The Ellipsoid Points algorithm generates a grid of points that are contained in an ellipsoid with a | |
| − | + | given spacing. To increase the efficiency of the algorithm the points from one of the eight quadrants are generated and the corresponding | |
| − | + | points from the other seven quadrants are calculated by including all the possible combinations of positive and negative values. Another | |
| − | + | function in this module sets up a set of points for Dijkstra’s Algorithm by checking to see if the point is an obstacle, setting the points | |
| − | + | to unvisited, setting the distance to be infinite and the previous node to be an empty list.</p> | |
| − | + | <figure> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/b/bd/T--Queens_Canada--RedPointsEllipsoid.jpg" alt='redpoints'/> | |
| − | + | <figcaption>The red points in this image represent the point grid that is developed for later use in the path finding algorithm.</figcaption> | |
| − | + | </figure> | |
| − | + | ||
| − | + | <h6>Dijkstra R3</h6> | |
| − | + | <p>This module implements an extended version of Dijkstra’s algorithm that allows for the shortest path through a | |
| − | + | set of points in R3. The algorithm implements a heuristic approach using the Euclidean distance of the current point to the end of the | |
| − | + | proposed linker site.</p> | |
| − | + | ||
</div> | </div> | ||
<!-- INFORMATION TO BE RELOCATED | <!-- INFORMATION TO BE RELOCATED | ||
| − | <div style="width: | + | <div style="width:70%;margin-left:15%"> |
| − | + | <h2>Eric's Latex files (to insert)</h2> | |
| − | + | <p>We determined that the linkers between the GR-LBD and the inteins halves would need to be flexible as | |
| − | + | there is no direct path connecting the termini and they would require the flexibility to meet each other | |
| − | + | and produces a trans-splicing event. To determine which linker would be ideal we ran simulations of | |
| − | + | Root-mean-square deviation of atomic positions to determine optimal linker length and composition. | |
| − | + | Firstly, we chose various linker designs inspired from previous work [a,b], then the entropy equations | |
| − | + | were run on the Queen’s University Computer Cluster for seven days by Dr. Campbell. | |
| − | + | <ol type="a"> | |
| − | + | <li><a href="http://parts.igem.org/Protein_domains/Linker">http://parts.igem.org/Protein_domains/Linker</a></li> | |
| − | + | <li><a href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3726540/">https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3726540/</a></li> | |
| − | + | </ol> | |
| − | + | </p> | |
| − | + | <p>Another aspect of our project we sought to model was which pacifier design would allow for optimal detection of the luminescence signal. | |
| − | + | Eric modelled this through a MATLAB simulation of the “Narrow escape problem”.... | |
| − | + | </p> | |
| − | + | <p>Lastly, the analysis was modeled by… | |
| − | + | </p> | |
| − | + | ||
| − | + | <h5>Molecular Dynamics</h5> | |
| − | + | <p>To model our target system and to get a more realistic view on how the linkers would perform, the constructs were modelled on PyMOL with some | |
| − | + | of the initial linker designs. These models were then sent to the cluster computers here at Queen’s University to undergo a molecular dynamics | |
| − | + | simulation. Dynamic modelling of molecular constructs allows for the molecule to simulate how it would vibrate in space. | |
| − | + | </p> | |
| − | + | <figure> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/6/62/T--Queens_Canada--PyMOLEstrogenLinker.jpg" alt='glucocorticoid'/> | |
| − | + | <figcaption>Linked glucocorticoid receptor and recA intein halves using the linkers from the Estrogen intein system paper. | |
| − | + | </figcaption> | |
| − | + | </figure><br> | |
| − | + | <p>This allows for qualification of the linkers’ length and flexibility. By examining the proximity between the two halves of the intein we can | |
| − | + | determine if an association event is possible. If the linkers do not appear to reach or if there appears to be excess length causing the halves | |
| − | + | of the inteins to overshoot each other the linker can be qualified as either too short or too long. The flexibility can be qualified by watching | |
| − | + | to note how easily and often the two halves of the intein are able to come close to each other. If the halves of the intein are easily able to | |
| − | + | reach each other and do so more than once this indicates that the linker is probably at an appropriate flexibility for our purposes. If the halves | |
| − | + | easily reach each other but also flail around more than expected the linkers are likely too flexible. Conversely if the halves do not easily come | |
| − | + | into proximity than the linker is likely too inflexible. | |
| − | + | </p> | |
| − | --> | + | --> |
<footer style="background-color: #212121;height:90px "> | <footer style="background-color: #212121;height:90px "> | ||
<div class="container"> | <div class="container"> | ||
Revision as of 23:08, 18 September 2018
Linker Software Summary
Summary
The software being developed is designed to connect two selected points on the molecule by finding the shortest path that connects them without interfering with the rest of the molecule. This is done by first finding the shortest path at a specified resolution using an extended version of Dijkstra’s Algorithm. After the shortest path has been found linkers are designed to suit the path found. The method used to generate the linkers has yet to been determined. This software will be usable through the PyMOL software to account for visual representation and interaction with the molecule of choice.
File Description
Main File
The main file interfaces with PyMOL allowing for user inputs to be used in the linker software. In addition, this file processes the inputs and manages all the function calls that coordinate the creation of the linkers.

Ellipsoid Fit
The ellipsoid fit module finds the tightest fitting ellipsoid to a set of points that represent the atoms that compose the molecule. This is done using an algorithm that is based on the Khachiyan Algorithm. This module returns the center, radii and rotation of the ellipsoid that encloses the points and allows for the plotting of the ellipsoid as a mesh graph.
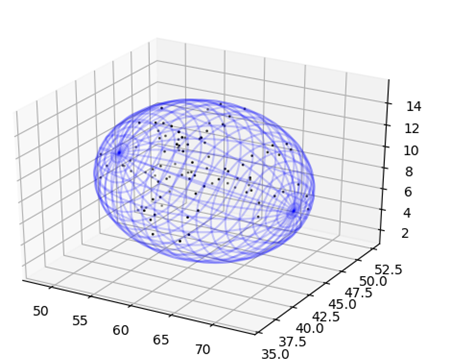
Ellipsoid Points
The Ellipsoid Points algorithm generates a grid of points that are contained in an ellipsoid with a given spacing. To increase the efficiency of the algorithm the points from one of the eight quadrants are generated and the corresponding points from the other seven quadrants are calculated by including all the possible combinations of positive and negative values. Another function in this module sets up a set of points for Dijkstra’s Algorithm by checking to see if the point is an obstacle, setting the points to unvisited, setting the distance to be infinite and the previous node to be an empty list.

Dijkstra R3
This module implements an extended version of Dijkstra’s algorithm that allows for the shortest path through a set of points in R3. The algorithm implements a heuristic approach using the Euclidean distance of the current point to the end of the proposed linker site.
