Met differ
Data format
We design a data format, .met format, to simply describe a metabolic network. .met format is easy to read and write.
If a line starts with ‘##’, this line indicates a new subgraph. Follows the ‘##’ is the name of the subgraph. And the next lines are this subgraph.
If a line starts with ‘#’, this line is a metabolite. In this line, there should be metabolite ID, name and other information like SMILES.
If a line starts without any symbol, it indicates a reaction. Reaction ID, reactors and products should be included. And if exists, enzyme information should be included as well.
If a line starts with ‘##’, this line indicates a new subgraph. Follows the ‘##’ is the name of the subgraph. And the next lines are this subgraph.
If a line starts with ‘#’, this line is a metabolite. In this line, there should be metabolite ID, name and other information like SMILES.
If a line starts without any symbol, it indicates a reaction. Reaction ID, reactors and products should be included. And if exists, enzyme information should be included as well.

Algorithm
In this part, we use merge-and-mine method. First, we merge the pathway graph and the network graph into one graph called align graph, according to the similarity coefficient of nodes. Then the nodes in the align graph are lined according to topology structure. Finally, we search for maximal connected subgraph as the alignment result.
Similarity coefficient
When we align the pathway to the network, similarity should be qualified. We use similarity coefficient to value the similarity between the pathway and the network. Here we consider the similarity from two aspects, node and topology structure.

NodeThere are two kinds of nodes in the metabolic network, metabolites and reactions. We consider them separately.
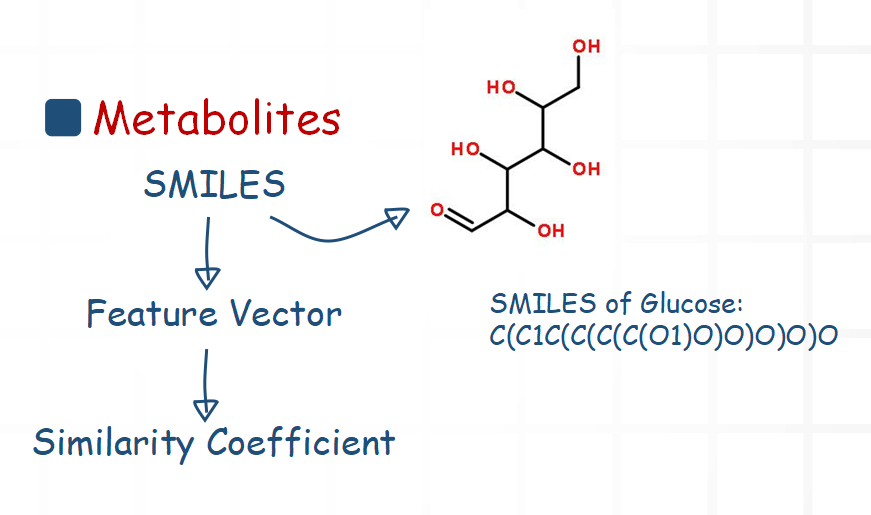
For metabolites, we mainly compare their structure information. Chemical structural formula can be described with SMILES. So we use a software package to extract a feature vector from the SMILES. Then we calculate the similarity coefficient of two feature vectors.

For reactions, we mainly consider the enzyme information. For each enzyme has an EC number, we can simply regularize the EC number to be the enzyme’s feature vector. For those reactions without an EC number, we use their reactors’ and products’ information to be the reaction’s information.

