(Created page with "<html lang="en"> <head> <meta charset="utf-8"> <title>HillSide Multi purpose HTML5 Template</title> <meta name="viewport" content="width=device-width, initial-scale=1.0" /> <m...") |
|||
| Line 242: | Line 242: | ||
<i class="icon-info-blocks material-icons hidden-xs"><img class="img-responsive" src="https://static.igem.org/mediawiki/2018/8/8d/T--HUST-China--2018-lanzao01.png"></i> | <i class="icon-info-blocks material-icons hidden-xs"><img class="img-responsive" src="https://static.igem.org/mediawiki/2018/8/8d/T--HUST-China--2018-lanzao01.png"></i> | ||
<div class="info-blocks-in" style="background-color: #ffffff; border: 1px solid #eeeeee;border-radius:5px;"> | <div class="info-blocks-in" style="background-color: #ffffff; border: 1px solid #eeeeee;border-radius:5px;"> | ||
| − | <h3>1. | + | <h3>1.<i>Synechocystis</i> </h3> |
| − | <div class="col-md-12" > <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/ | + | <div class="col-md-12" > <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/d/df/T--HUST-China--2018-model-new-c001.png"></div> |
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <p>These are the main differential equations of the | + | <p>These are the main differential equations of the Synechocystis part of the model. The parameters and variables above will be introduced below.</p> |
</div> | </div> | ||
<div class="col-md-10" > | <div class="col-md-10" > | ||
| − | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/ | + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/8/8c/T--HUST-China--2018-model-new-c002.png"> |
</div> | </div> | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <p>These functions calculate the growth rate of | + | <p>These functions calculate the growth rate of Synechocystis. The two functions correspond to different concentrations of CO<sub>2</sub> in the solution.<img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/3/36/T--HUST-China--2018-model-new-c003.png">and<img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/b/bf/T--HUST-China--2018-model-new-c004.png">make the growth rate decline when the concentration of CO<sub>2</sub> in the solution is too high or too low. (David et al. 2015)<sup>[1]</sup>. From the same reference we get the relationship between the light intensity and the growth rate. The function<img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/d/d8/T--HUST-China--2018-model-new-c005.png">is from another reference(XIONG et al. 2012) <sup>[2]</sup>. The parameter <img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/0/0c/T--HUST-China--2018-model-new-c006.png"> the decline of the growth rate caused by gene editing. This parameter is introduced to make the growth model of wild Synechocystis fit to the growth of edited ones in the reference (Henrike et al. 2010)<sup>[3]</sup>.</p> |
</div> | </div> | ||
<div class="col-md-6" > | <div class="col-md-6" > | ||
| − | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/ | + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/c/c4/T--HUST-China--2018-model-new-c007.png"> |
</div> | </div> | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <p>This stands for the CO<sub>2</sub> consumed by each unit of | + | <p>This stands for the CO<sub>2</sub> consumed by each unit of Synechocystis.<img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/c/cc/T--HUST-China--2018-model-new-c008.png">is used for growth,<img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/1/1d/T--HUST-China--2018-model-new-c009.png">is used for lactate producing, and <img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/c/cb/T--HUST-China--2018-model-new-c010.png"> is used for sustaining its life.</p> |
</div> | </div> | ||
<div class="col-md-3" > | <div class="col-md-3" > | ||
| − | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/ | + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/2/26/T--HUST-China--2018-model-new-c011.png"> |
</div> | </div> | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <p>The O<sub>2</sub> consuming is calculated by | + | <p>The O<sub>2</sub> consuming is calculated by <img style="vertical-align:middle" src="https://static.igem.org/mediawiki/2018/8/89/T--HUST-China--2018-model-new-c013.png"> because the photosynthesis and respiration in Synechocystis have the same stoichiometric ratio.</p> |
</div> | </div> | ||
<div class="col-md-6" > | <div class="col-md-6" > | ||
| − | + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/9/96/T--HUST-China--2018-model-new-c012.png"> | |
</div> | </div> | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
| − | <p>This function shows the inhibit of CO<sub>2</sub> to the metabolism of | + | <p>This function shows the inhibit of CO<sub>2</sub> to the metabolism of Synechocystis. The lack of carbon source will not only effect the growth rate of the bacteria, but also reduce the produce rate of lactate.</p> |
</div> | </div> | ||
Revision as of 17:02, 17 October 2018
Comparison_between_PSB
1. Abstract
Modelling is a powerful tool in synthetic biology that allows us to get a deeper understanding of our system. In order to see whether our system can work and how our system will work, we build this model to simulate our system. This model shows us the details of our system and give our intelligent device software data so that we can change the environment to increase the efficiency and stability of Optopia.
2. Overview
functions

1.Synechocystis

These are the main differential equations of the Synechocystis part of the model. The parameters and variables above will be introduced below.

These functions calculate the growth rate of Synechocystis. The two functions correspond to different concentrations of CO2 in the solution. and
and make the growth rate decline when the concentration of CO2 in the solution is too high or too low. (David et al. 2015)[1]. From the same reference we get the relationship between the light intensity and the growth rate. The function
make the growth rate decline when the concentration of CO2 in the solution is too high or too low. (David et al. 2015)[1]. From the same reference we get the relationship between the light intensity and the growth rate. The function is from another reference(XIONG et al. 2012) [2]. The parameter
is from another reference(XIONG et al. 2012) [2]. The parameter  the decline of the growth rate caused by gene editing. This parameter is introduced to make the growth model of wild Synechocystis fit to the growth of edited ones in the reference (Henrike et al. 2010)[3].
the decline of the growth rate caused by gene editing. This parameter is introduced to make the growth model of wild Synechocystis fit to the growth of edited ones in the reference (Henrike et al. 2010)[3].

This stands for the CO2 consumed by each unit of Synechocystis. is used for growth,
is used for growth, is used for lactate producing, and
is used for lactate producing, and  is used for sustaining its life.
is used for sustaining its life.

The O2 consuming is calculated by  because the photosynthesis and respiration in Synechocystis have the same stoichiometric ratio.
because the photosynthesis and respiration in Synechocystis have the same stoichiometric ratio.

This function shows the inhibit of CO2 to the metabolism of Synechocystis. The lack of carbon source will not only effect the growth rate of the bacteria, but also reduce the produce rate of lactate.

2.Rhodopseudomonas palustris

These are the main differential equations about the modeling part of the Rhodopseudomonas palustris (abbreviated as “Rps”). Through these differential equations, we can calculate the concentrations of Rps, carbon dioxide and lactate. The parameters and variables above will be introduced below.This part is similar to that of Cyanobacteria, because in our experiment, they are both used to provide lactate to Shewanella and have similar genetic modification.

The function calculates the growth rate of Rhodopseudomonas palustris. The function corresponds to different concentrations of CO2 in the solution. makes the growth rate decline when the concentration of CO2 in the solution is too high. This is gotten from the reference (Can. J. Chem.et al. 2015 ).
makes the growth rate decline when the concentration of CO2 in the solution is too high. This is gotten from the reference (Can. J. Chem.et al. 2015 ).
From the same reference we get the relationship between the light intensity and the growth rate. The function

is from another reference (Xiong et al. 2012). The parameter kdeclineis the decline of the growth rate caused by gene editing. This parameter is introduced to make the growth model of wild Rhodopseudomonas palustris fit to the growth of edited ones in the reference ( Henrike et al. 2012 ).

This stands for the CO2 consumed by each unit of Rhodopseudomonas palustris.  is used for growth,
is used for growth,  is used for lactate producing, and mCO2,Rpsis used for sustaining its life.
is used for lactate producing, and mCO2,Rpsis used for sustaining its life.

This function shows the inhibit of CO2 to the metabolism of Rhodopseudomonas palustris. The lack of carbon source will not only impact the growth rate of the bacteria, but also reduce the production rate of lactate.

3. Shewanella oneidensis
In generate, the three elements urgently needed to be modeled in Shewanella are the changes in biomass (Dry Weight, g/L), electricity production (mV), and lactate content (g/L) over time. Once the Shewanella modeling is completed, we only need to combine the model of Shewanella with the model of Cyanobacteria or Rhodopseudomonas palustris to determine which one is better to facilitate electricity produce. The process of deduction will write blow:
First, we need to simulate biomass function. Our biomass function is based on monod equation:

In this function, μShewa is specific growth rate of biomass.
Because there are two important growth factors in our model: lactate and oxygen content, we need some modifying tasks in this model. Noticing that the concentration of oxygen and lactate are both promoting biomass growth, with the inspiration of monod equation, we take the two factors into consideration so the function changes to:

Shewanella oneidensis MR-1 prefers to use lactate as its carbon source since the amount of lactate-based biomass is more than acetate-based biomass or pyruvate-based biomass. Dld and lldEFG are D- and L-lactate dehydrogenase enzymes, which is the first step of utilizing lactate. To make the use of lactate more efficiently, we overexpress four genes: dld, lldE, lldF, lldG.[10]
①. dld: dld refers to FAD-dependent D-lactate dehydrogenase which could catalyze D-lactate’s transformation into pyruvate.
②. lldEFG: They could encode a L-lactate dehydrogenase complex which could catalyze D-lactate’s transformation into pyruvate.
To ensure that the genes would be expressed efficiently, we add a promoter before lldEFG:
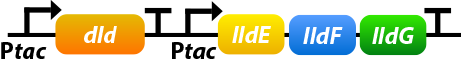
NADH is a significant part of extracellular electron transfer(EET) as it could carry electron. Strenghthening the regeneration of NADH would make EET more efficiently.
To achieve this goal, we overexpress these four genes: gapA2, mdh, pflB, fdh. [11]
①. gapA: It encodes glyceraldehyde-3-phosphate dehydrogenase which could transform 3- phosphoglyceraldehyde into 1,3- diphosphoglycerate.
②. mdh: It encodes NAD dependent malate dehydrogenase which transforms malate into pyruvate
③. pflB: It encodes pyruvate formate-lyase to transform pyruvate into Acetyl-CoA.
④. fdh: It encodes formate dehydrogenase to transform formate into CO2..
Also, to ensure that the genes would be expressed efficiently, we add an promoter before pflB and fdh:

Part3: Whole design

Design of MFC
We have designed a bipolar chamber MFC this year. Proton exchange membrane divided it into anode chamber and cathode chamber. Anode chamber containing S.oneidensis, nutrient substance(LB、lactate ) or other electrical producing microbes were sealed to prevent the entry of external oxygen. Considering safety and oxidation-reduction potential, we put ferric chloride solution in cathode chamber so that S.oneidensis can transfer electrons outside of their membranes by electron transport chain. Then electrons will reduce ferric ion into ferrous through carbon cloth and produce electricity.We recorded open circuit voltage curve and load voltage curve of MFCs in each different systems. Also, we have measured the biomass of each system in order to ensure whether the improved electricity could be attributed to more attached Shewanella cells on the anodes or the higher electroactivity of single cell.[11]

Co-culture
Obviously, the ecological relationship between microorganisms is very complex. There is not only the competition between them for the nutrient, but also the regulation of metabolites among them including induction, transgenosis and synergistic metabolism. Besides, it has been found that the co-culture of microorganisms can improve the electric efficiency of Microbial Fuel Cell under certain conditions.
Metabolites exchange is a common relationship in co-culturing. Therefore, we have designed a clear microbial metabolic pathway to achieve the conversion from light to electricity as well as used more potential symbiotic relationships between the flora to help improve the electricity production efficiency of MFC.
By consulting literature, we found two kinds of microorganisms——Cyanobacteria and Rhodopseudomonas palustris, both of which can utilize light energy and provide lactate to S.oneidensis after doing molecular construction.
In order to provide a basic growth environment, we mix the culture medium of different strains.(Please refer to our protocol section for the composition of the mediums.)
Synechocystis PCC6803
Lactate produced by Synechocystis PCC6803 can be used as the optimal carbon source for Shewanella. At the same time, acetate produced by Shewanella can be used as the organic carbon source of Synechocystis PCC6803 to increase the lactate production. And the metabolite exchange of Synechocystis PCC6803 and Shewanella is the basis for our photoautotrophic MFC.[12].
Rhodopseudomonas palustris
We attempted to engineer Rhodopseudomonas palustris by synthetic biology to achieve the same or a better function of Synechococcus elongatus.
In the preliminary experiment, we found that there may be more potential interactions in the co-culture of Rhodopseudomonas palustris and Shewanella, which can greatly improve the coulombic efficiency of our MFC (please refer our results section for more detials). This is an unexpected surprise for us, which improve to our confidence in the success of the project.
Reference
[1]Henrike Niederholtmeyer, Bernd T. Wolfstädter, David F. Savage, Pamela A. Silver, Jeffrey C. Way. Engineering Cyanobacteria to Synthesize and Export Hydrophilic Products. Applied and Environmental Microbiology. 2010, 76(11): 3462-3466.
[2] 《沼泽红假单胞菌的研究进展及其应用》王跃先、刘德海
[3] 周茂洪 赵肖为 吴雪昌《光合细菌沼泽红假单胞菌同化磷能力的研究》 《科技通报》2002年 第2期 142-146页
[4] KEGG, https://www.genome.jp/kegg/pathway.html
[5] Cloning and sequence analysis of the gene encoding Lactococcus lactis malolactic enzyme: relationships with malic enzymes FEMS Microbiol Lett. 1994 Feb 1;116(1):79-86
[6] Fine tuning the transcription of ldhA for d-lactate production August 2012, Volume 39, Issue 8, pp 1209–1217
[7] Transport of L-Lactate, D-Lactate, and Glycolate by the LldP and GlcA Membrane Carriers of Escherichia coli Volume 290, Issue 2, 18 January 2002, Pages 824-829
[8] Enhancement of Hydrogen Production and Carbon Fixation in Purple Nonsulfur Bacterium Bacterium by Synthetic Biology Shou-Chen Lo
[9] Jcat, http://www.jcat.de/#opennewwindow
[10] Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization PNAS February 24, 2009 106 (8) 2874-2879
[11] Modular Engineering Intracellular NADH Regeneration Boosts Extracellular Electron Transfer of Shewanella oneidensis MR‑1 ACS Synth. Biol. 2018, 7, 885−895
[12]Varman A. M., Yu Y., You L. & Tang Y. J. Photoautotrophic production of d-lactate in an engineered cyanobacterium. Microb. Cell Fact. 12, 117 (2013).



