|
|
| (6 intermediate revisions by 2 users not shown) |
| Line 2: |
Line 2: |
| | {{Marburg}} | | {{Marburg}} |
| | <html> | | <html> |
| − | <div class="titleWrapper"><div class="titleBackground" style="background-image:none"></div><div class="title">Improve</div></div> | + | <div class="titleWrapper"><div class="titleBackground" style="background-image:url(https://static.igem.org/mediawiki/2018/5/54/T--Marburg--header_improve.jpg)"></div><div class="title">Improve</div></div> |
| | <article> | | <article> |
| | <style> | | <style> |
| Line 47: |
Line 47: |
| | <p> | | <p> |
| | <br /> | | <br /> |
| − | Cloning is the daily bread of molecular biologists. Almost all synbio projects start with weeks or months of cloning, a time which is the most frustrating and simultaneously least rewarding period of a project. Being able to distinguish between the native vector and the successfully assembled plasmid clearly while picking can save a lot of time and work. During the last decades, many approaches have been established to help distinguishing between false and correctly assembled plasmids. In our project, we created the Marburg Collection, a novel golden-gate-based cloning toolbox. This toolbox consists of LVL0 parts stored in a
| + | Cloning is the daily bread of molecular biologists. Almost all synbio projects start with weeks or months of cloning, a time which is the most frustrating and simultaneously least rewarding period of a project. Being able to distinguish between the native vector and the successfully assembled plasmid clearly while picking can save a lot of time and work. During the last decades, many approaches have been established to help distinguishing between false and correctly assembled plasmids. In our project, we created the Marburg Collection, a novel golden-gate-based cloning toolbox. This toolbox consists of LVL0 parts stored in a |
| | <a href=" http://parts.igem.org/Part:pSB1C3 "> | | <a href=" http://parts.igem.org/Part:pSB1C3 "> |
| | <abbr title=" Link to the iGEM part registry "> | | <abbr title=" Link to the iGEM part registry "> |
| Line 53: |
Line 53: |
| | </abbr> | | </abbr> |
| | </a> | | </a> |
| − | derivat. As many plasmids had to be created, we desperately needed a simple, fast and cheap way to reliably select correct colonies even for inefficient clonings. | + | derivat. |
| − | </p>
| + | As many plasmids had to be created, we desperately needed a simple, fast and cheap way to reliably select correct colonies even for inefficient clonings. |
| − | <p>
| + | |
| − | Here, we present the improvement of an iGEM-part for (a mostly faster,) easier and cheaper selection which suits our needs.
| + | |
| | </p> | | </p> |
| | + | |
| | + | |
| | <p> | | <p> |
| | + | Here, we present the improvement of an iGEM-part for (a mostly faster,) easier and cheaper selection which suits our needs. |
| | + | </p><figure style="display:block; margin:0 auto 0 auto; width: 50%;"> |
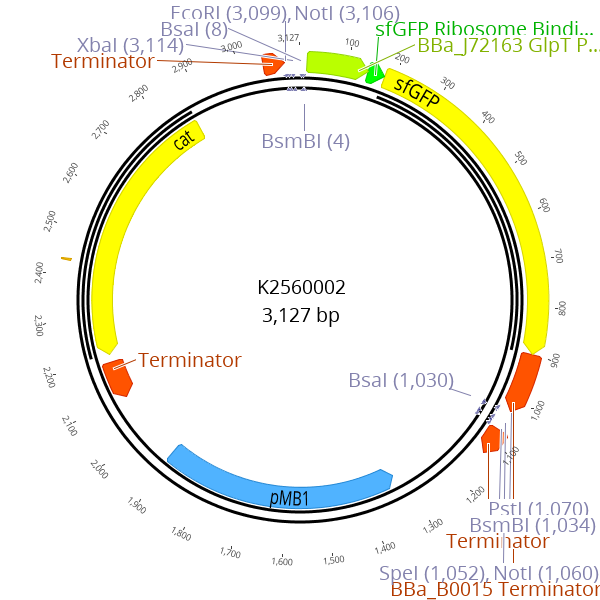
| | + | <img src=" https://static.igem.org/mediawiki/2018/8/85/T--Marburg---K2560002.png"> |
| | + | <figcaption><b> Figure 1: Plasmidmap of BBa_K2560002 </b><br>Our improved part BBa_K2560002 is a derivate of the BBa_P10500 containing sfGFP drop out.</figcaption> |
| | + | </figure> |
| | + | |
| | + | <p> |
| | iGEM provides the universal acceptor plasmid | | iGEM provides the universal acceptor plasmid |
| | <a href=" http://parts.igem.org/Part:BBa_P10500 "> | | <a href=" http://parts.igem.org/Part:BBa_P10500 "> |
| Line 65: |
Line 72: |
| | </a> | | </a> |
| | for creating new PhytoBricks. This plasmid contained the <i>lacZ-α</i> part in the cloning sites for blue-white screening. The <i>lacZ</i>-gene encodes the β-galactosidase, which catalyzes the hydrolysis of the glycosidic bond of β-galactopyranosides like D-lactose. Blue-white screening is based on alpha-complementation, where the <i>α</i>- subunit (C-terminal section) and <i>ω</i>-subunit (N-terminal section) of the β-galactosidase (both non-functional peptides) are able to reconstitute a functional enzyme. By the addition of the two D-lactose Analogues Xgal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) and IPTG (Isopropyl ß-D-1-thiogalactopyranoside) to the plates, colonies with successful cloning products thereby no <i>lacZ-α</i> will have the typical white colour while the unsuccessful ones precipitate the blue-coloured product resulting in blue colonies. | | for creating new PhytoBricks. This plasmid contained the <i>lacZ-α</i> part in the cloning sites for blue-white screening. The <i>lacZ</i>-gene encodes the β-galactosidase, which catalyzes the hydrolysis of the glycosidic bond of β-galactopyranosides like D-lactose. Blue-white screening is based on alpha-complementation, where the <i>α</i>- subunit (C-terminal section) and <i>ω</i>-subunit (N-terminal section) of the β-galactosidase (both non-functional peptides) are able to reconstitute a functional enzyme. By the addition of the two D-lactose Analogues Xgal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) and IPTG (Isopropyl ß-D-1-thiogalactopyranoside) to the plates, colonies with successful cloning products thereby no <i>lacZ-α</i> will have the typical white colour while the unsuccessful ones precipitate the blue-coloured product resulting in blue colonies. |
| − | <b> For further reading: jana design/results </b> </p> | + | For further reading, <a href="https://2018.igem.org/Team:Marburg/Results"><abbr title="Link to Results">click here. </abbr></a> <br> However, this kind of selection requires the use of specific strains which possess the <i>lacZ-ω</i> but neither the native <i>lacZ</i> nor the <i>lacZ-α</i> fragment. As an example, the |
| − | | + | |
| − | <p>
| + | |
| − | However, this kind of selection requires the use of specific strains which possess the <i>lacZ-ω</i> but neither the native <i>lacZ</i> nor the <i>lacZ-α</i> fragment. As an example, the
| + | |
| | <a href=" https://www.neb.com/-/media/catalog/datacards-or-manuals/c2984datasheet-lot1131402.pdf "> | | <a href=" https://www.neb.com/-/media/catalog/datacards-or-manuals/c2984datasheet-lot1131402.pdf "> |
| | <abbr title="""> | | <abbr title="""> |
| Line 77: |
Line 81: |
| | BBa_P10500</abbr> | | BBa_P10500</abbr> |
| | </a> | | </a> |
| − | in <i>V. natriegens</i> shows no difference between colonies containing the <i>lacZ-α</i> dropout and those without. <b> For further reading about the <i>lac</i> operon of <i>V. natriegens</i>, click here. </b></p> | + | in <i>V. natriegens</i> shows no difference between colonies containing the <i>lacZ-α</i> dropout and those without. For further reading about the <i>lac</i> operon of <i>V. natriegens</i>, <a href="https://2018.igem.org/Team:Marburg/Design"><abbr title="Link to Design">click here. </b></p></abbr></a></p> |
| | <p> | | <p> |
| | To overcome this limitation and to enable fast and reliable cloning with <i>V. natriegens</i>, we decided to establish a new visualization method. Our improved part, namely | | To overcome this limitation and to enable fast and reliable cloning with <i>V. natriegens</i>, we decided to establish a new visualization method. Our improved part, namely |
| Line 90: |
Line 94: |
| | <dfn data-info=" superfolded green fluorescent protein "> | | <dfn data-info=" superfolded green fluorescent protein "> |
| | sfGFP</dfn> | | sfGFP</dfn> |
| − | dropout as fluorescent selection marker. In the pictures xxx on the right, you can see the wild type <i>V. natriegens</i> in comparison with the iGEM | + | dropout as fluorescent selection marker. |
| | + | |
| | + | <br> |
| | + | |
| | + | |
| | + | |
| | + | In the pictures 2, you can see the wild type <i>V. natriegens</i> in comparison with the iGEM |
| | <a href=" http://parts.igem.org/Part:BBa_P10500 "> | | <a href=" http://parts.igem.org/Part:BBa_P10500 "> |
| | <abbr title=" Link to the iGEM part registry "> | | <abbr title=" Link to the iGEM part registry "> |
| Line 106: |
Line 116: |
| | BBa_K2560002</abbr></a> part leads to a strong visual distinction to colonies which do not possess the | | BBa_K2560002</abbr></a> part leads to a strong visual distinction to colonies which do not possess the |
| | <dfn data-info=" superfolded green fluorescent protein "> | | <dfn data-info=" superfolded green fluorescent protein "> |
| − | sfGFP</dfn>. In this way, we created a part for universal LVL0 cloning with a improved selection without the need of additional supplements like Xgal or IPTG. </p> | + | sfGFP</dfn>. In this way, we created a part for universal LVL0 cloning with a improved selection without the need of additional supplements like Xgal or IPTG. </p> Our part not only suits for <i>V. natriegens</i> but is convenient for the frequently used cloning host <i>E. coli</i>. As it can be seen in the picture 3, <i>E. coli</i> containing the |
| − | <p>
| + | |
| − | Our part not only suits for <i>V. natriegens</i> but is convenient for the frequently used cloning host <i>E. coli</i>. As it can be seen in the picture xxx, <i>E. coli</i> containing the
| + | |
| | <dfn data-info=" superfolded green fluorescent protein "> | | <dfn data-info=" superfolded green fluorescent protein "> |
| | sfGFP</dfn> possess a considerable strong green colour even without the use of UV light and is just or even more distinguishable from the wild type as the <i>lacZ</i> containing blue colonies. By using our improved part | | sfGFP</dfn> possess a considerable strong green colour even without the use of UV light and is just or even more distinguishable from the wild type as the <i>lacZ</i> containing blue colonies. By using our improved part |
| Line 123: |
Line 131: |
| | <abbr title=" link to shop "> | | <abbr title=" link to shop "> |
| | IPTG</abbr></a> | | IPTG</abbr></a> |
| − | costs about 80 dollar. </p> | + | costs about 80 dollar. |
| − | <p>By using our improved part for cloning, the detection of successfully ligated clones from non is feasible for strains which are not compatible with blue-white screening and therefor becomes more universal, faster and cheaper than before. | + | |
| | + | <style> |
| | + | .imageContainer { |
| | + | display: flex; |
| | + | justify-content: space-around; |
| | + | align-items: center; |
| | + | } |
| | + | .imageContainer img { |
| | + | width: 80%; |
| | + | } |
| | + | </style> |
| | + | |
| | + | <div class="imageContainer"> |
| | + | <figure> |
| | + | <img src=" https://static.igem.org/mediawiki/2018/8/80/T--Marburg--V.natriegens.png "/> |
| | + | |
| | + | <figcaption><b> Figure 2: <i>V. natriegens</i> </b><br> A: Colonies of wild type, B: white colonies containing BBa_P10500 and C: colonies with BBa_K2560002 resulting in green colonies. </figcaption> |
| | + | </figure> |
| | + | </img> |
| | + | <figure> |
| | + | <img src=" https://static.igem.org/mediawiki/2018/b/b3/T--Marburg--E.coli.png "/> <figcaption><b> Figure 3: <i>E. coli</i> </b><br> A: Colonies of wild type, B: blue colonies containing BBa_P10500 and C: colonies with BBa_K2560002 resulting in green colonies. </figcaption> |
| | + | </figure> |
| | + | </img> |
| | + | |
| | + | </div> |
| | + | </p> |
| | + | <p> <br>By using our improved part for cloning, the detection of successfully ligated clones from non is feasible for strains which are not compatible with blue-white screening and therefor becomes more universal, faster and cheaper than before. |
| | </p> | | </p> |
| | + | </body> |
| | + | |
| | + | </article> |
| | + | </html> |
| | + | |
| | + | {{Marburg/footer}} |