| Line 222: | Line 222: | ||
<div class="row section-header has-bottom-sep" data-aos="fade-up"> | <div class="row section-header has-bottom-sep" data-aos="fade-up"> | ||
<div class="col-full"> | <div class="col-full"> | ||
| − | <h3 class="subhead subhead--dark">Naringenin Operon</h3> | + | <h3 class="subhead subhead--dark">Improved Naringenin Operon based on Pathway Modelling</h3> |
<h1 class="display-1 display-1--light">DESIGN</h1> | <h1 class="display-1 display-1--light">DESIGN</h1> | ||
</div> | </div> | ||
| Line 229: | Line 229: | ||
<div class="row about-desc" data-aos="fade-up"> | <div class="row about-desc" data-aos="fade-up"> | ||
<div class="col-full"> | <div class="col-full"> | ||
| − | <p class="about-para"> | + | <p class="about-para"> Results of the modelling showed that weaker expression of the first two genes and 1000 fold stronger expression of the last two genes would reduce the build-up of malonyl CoA and optimise naringenin synthesis. Therefore we found two synthetic promoters: BG28 and BG51 and two E. coli his operon terminator. These will allow enhanced naringenin production in future experiments.</p> |
</div> | </div> | ||
Revision as of 14:13, 9 October 2018
Alternative Roots
Design
================================================== -->
Hydroponics
DESIGN
Once the project idea was finalised, the team began looking for cheap, efficient and standardised methods for growing plants in iGEM. The hope was that such an item existed that would meet these specifications as well as being a closed container to prevent contamination and also providing a high throughput of plants. It was soon established that such an item did not exist to meet our specifications. Therefore, to combat this issue, it was decided that the best way forward would be to design our own hydroponics system. This would allow us to grow large amounts of Arabidopsis in a controlled setting for the purposes of our project. Several team members were assigned to this “sub-project”.
Before getting hands-on in building the system, the team as a whole established a few design parameters. For example, the system needed to be cheap and easy to build from scratch. This is so future iGEM teams are able to construct the system for their own needs and even build upon our design, as necessary. Additionally, the system must be versatile, open-source and easily adapted for various conditions such as light intensity and wavelength. By adopting such an open and adaptable design the intention is that the end-user is able to effortlessly match the system to their needs, without getting entangled in streams of code.
Several weeks were spent modifying the design until a design was found that met all the above criteria, the specifications of the design can be seen below.
UP TO
SEEDS CAN BE GROWN
IN HYDROPONICS
APPROXIMATELY
KWH OF POWER ANNUALLY
USED TO POWER SYSTEM
PROVIDES UP TO
LUX OF LIGHT
TO GROW SEEDS
CONTAINS
INDIVIDUALLY ADDRESSABLE
LOW-POWER LED'S
Naringenin Operon
DESIGN
In order to produce the flavonoid naringenin, construction of a naringenin operon that would be synthesised from the sequence in the iGEM parts registry to gblocks by IDT was required. This operon was based on that used by the TU Darmstadt 2014 iGEM team, allowing the production of nitrogen to attract nitrogen fixing bacteria. It contains the genes for four enzymes: 4 – Coumaryl ligase, Tyrosine ammonia lyase, Chalcone isomerase and Chalcone synthase. Each of these genes will contain a strong ribosome binding site. This construct will be under the control of a J23100 constitutive promoter to observe its expression in E. coli as a proof of concept. Once biosynthesis under the control of J23100 is achieved, the construct future experiments would test this under a T7 promoter in E. coli. Parts for biosynthesis in the final chassis organism, root-colonising Pseudomonas fluorescens, will be constructed in the plasmid backbone pSB1C3.
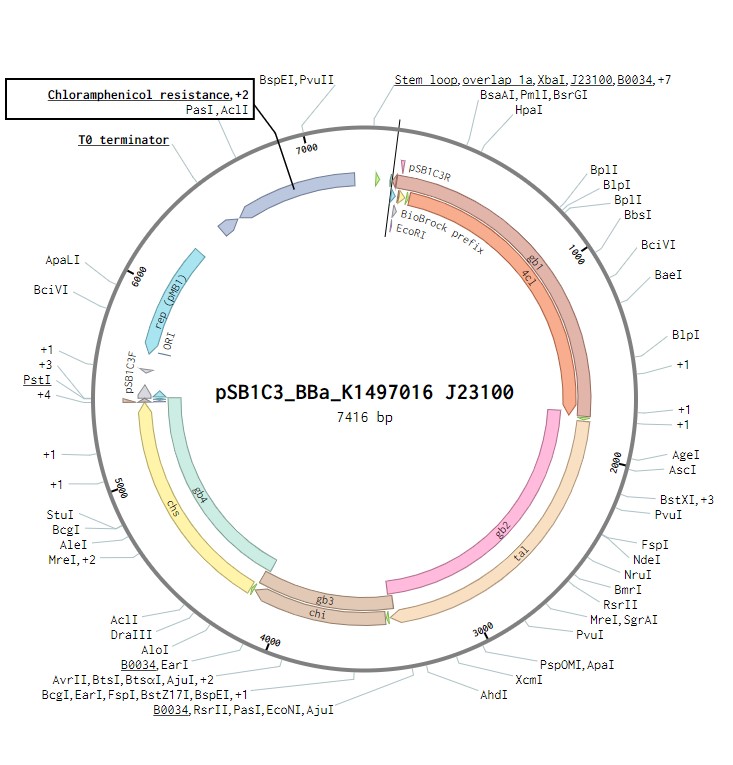
The naringenin biosynthetic operon under control of a J23100 promoter created in Benchling.

The naringenin biosynthetic operon under control of a T7 promoter created in Benchling.
UP TO
SEEDS CAN BE GROWN
IN HYDROPONICS
APPROXIMATELY
KWH OF POWER ANNUALLY
USED TO POWER SYSTEM
PROVIDES UP TO
LUX OF LIGHT
TO GROW SEEDS
CONTAINS
INDIVIDUALLY ADDRESSABLE
LOW-POWER LED'S
Improved Naringenin Operon based on Pathway Modelling
DESIGN
Results of the modelling showed that weaker expression of the first two genes and 1000 fold stronger expression of the last two genes would reduce the build-up of malonyl CoA and optimise naringenin synthesis. Therefore we found two synthetic promoters: BG28 and BG51 and two E. coli his operon terminator. These will allow enhanced naringenin production in future experiments.

