| Line 1: | Line 1: | ||
{{SBS_SH_112144}} | {{SBS_SH_112144}} | ||
| − | {{:Team:SBS_SH_112144/ | + | {{:Team:SBS_SH_112144/header2}} |
<html> | <html> | ||
<head> | <head> | ||
| Line 8: | Line 8: | ||
margin:0 auto; | margin:0 auto; | ||
text-align:left; | text-align:left; | ||
| + | } | ||
| + | .column{ | ||
| + | width:50%; | ||
} | } | ||
</style> | </style> | ||
| Line 13: | Line 16: | ||
<body> | <body> | ||
<div class="center"> | <div class="center"> | ||
| − | < | + | <h4>Molecular cloning</h4> |
<p>1. We were able to use PCR to successfully amplify the synthesized gene (gel unshown)</p> | <p>1. We were able to use PCR to successfully amplify the synthesized gene (gel unshown)</p> | ||
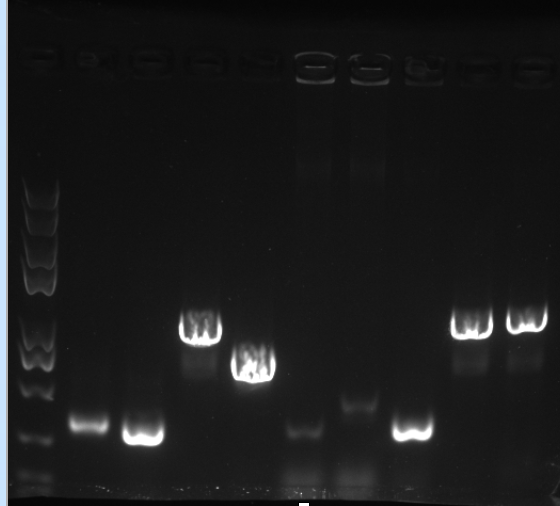
<p>2. Through homologous recombination using an exonuclease kit, we were able to fuse the cp-OS lysozyme 1 gene into the PET28A and PSB1C3 plasmid</p> | <p>2. Through homologous recombination using an exonuclease kit, we were able to fuse the cp-OS lysozyme 1 gene into the PET28A and PSB1C3 plasmid</p> | ||
<figure><center> | <figure><center> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/9/96/T--SBS_SH_112144--Cyanobacteria6.jpg" | + | <img src="https://static.igem.org/mediawiki/2018/9/96/T--SBS_SH_112144--Cyanobacteria6.jpg" style="width:500px; padding:20px 0px;"/> |
<figcaption>Gel electrophoresis:our lysozyme gene can be identified and amplified using the plasmid</figcaption></center> | <figcaption>Gel electrophoresis:our lysozyme gene can be identified and amplified using the plasmid</figcaption></center> | ||
</figure> | </figure> | ||
| − | + | <div style="clear:both; height:20px;"></div> | |
<p>3. We were able to cultivate the transformed BL21 E.coli and the addition of IPTG successfully triggered the expression of lysozyme. </p> | <p>3. We were able to cultivate the transformed BL21 E.coli and the addition of IPTG successfully triggered the expression of lysozyme. </p> | ||
| − | < | + | <h4>Protein Purification</h4> |
<p>By purifying the protein using nickel column and later cleaving the His tag using TEV enzyme, we obtained pure lysozyme testified by the clearly labeled gel below.</p> | <p>By purifying the protein using nickel column and later cleaving the His tag using TEV enzyme, we obtained pure lysozyme testified by the clearly labeled gel below.</p> | ||
<figure><center> | <figure><center> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/8/82/T--SBS_SH_112144--Cyanobacteria8_.jpg" | + | <img src="https://static.igem.org/mediawiki/2018/8/82/T--SBS_SH_112144--Cyanobacteria8_.jpg" style="width:500px; padding:20px 0px;" /> |
<figcaption>Protein Acrylamide Gel on the purified lysozyme</figcaption></center> | <figcaption>Protein Acrylamide Gel on the purified lysozyme</figcaption></center> | ||
</figure> | </figure> | ||
| + | <div style="clear:both; height:20px;"></div> | ||
| − | < | + | <h4>Function test</h4> |
<p>1. By applying the lysozyme to a system(300ml in a microfuge tube reacting under different temperature) with bugbuster, cyanobacteria, pH buffer and water, we are able to observe cleavage effect illustrated by the OD660nm green light absorptance across large temperature and pH range. And through a series of mathematical computation and <a href='https://2018.igem.org/Team:SBS_SH_112144/Model'>modeling</a >, we are able to identify that the ideal pH is around 6.9 and temperature is at 37 °C. </p> | <p>1. By applying the lysozyme to a system(300ml in a microfuge tube reacting under different temperature) with bugbuster, cyanobacteria, pH buffer and water, we are able to observe cleavage effect illustrated by the OD660nm green light absorptance across large temperature and pH range. And through a series of mathematical computation and <a href='https://2018.igem.org/Team:SBS_SH_112144/Model'>modeling</a >, we are able to identify that the ideal pH is around 6.9 and temperature is at 37 °C. </p> | ||
<figure><center> | <figure><center> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/f/fb/T--SBS_SH_112144--Cyanobacteria10_.jpg" | + | <img src="https://static.igem.org/mediawiki/2018/f/fb/T--SBS_SH_112144--Cyanobacteria10_.jpg" style="width:500px; padding:20px 0px;" /> |
<figcaption>Diagram we used to optimize pH and temperature</figcaption></center> | <figcaption>Diagram we used to optimize pH and temperature</figcaption></center> | ||
</figure> | </figure> | ||
| Line 42: | Line 46: | ||
<p>2. After finding the ideal pH and temperature for the lysozyme, we successfully applied these elements to a new set of function test with different protein concentration and reaction time, and we are also able to, through PSO algorithm and 3D modeling in matlab, find out the optimal reaction time being 0.5h( quite different from the paper) and the protein concentration being 18 ug/mL. </p> | <p>2. After finding the ideal pH and temperature for the lysozyme, we successfully applied these elements to a new set of function test with different protein concentration and reaction time, and we are also able to, through PSO algorithm and 3D modeling in matlab, find out the optimal reaction time being 0.5h( quite different from the paper) and the protein concentration being 18 ug/mL. </p> | ||
<figure><center> | <figure><center> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/5/5d/T--SBS_SH_112144--Cyanobacteria11_.jpg" | + | <img src="https://static.igem.org/mediawiki/2018/5/5d/T--SBS_SH_112144--Cyanobacteria11_.jpg" style="width:500px; padding:20px 0px;" /> |
<figcaption>Diagram we used to optimize enzyme concentration and reaction time</figcaption></center> | <figcaption>Diagram we used to optimize enzyme concentration and reaction time</figcaption></center> | ||
</figure> | </figure> | ||
| Line 48: | Line 52: | ||
<div class="row"> | <div class="row"> | ||
| − | <div class="column"> | + | <div class="column" style="float:left; text-align:center;"> |
| − | <img src= "https://static.igem.org/mediawiki/2018/4/4a/T--SBS_SH_112144--Cyanobacteria12_.jpg" | + | <img src= "https://static.igem.org/mediawiki/2018/4/4a/T--SBS_SH_112144--Cyanobacteria12_.jpg" style="width:500px; padding:20px 0px;"> |
</div> | </div> | ||
| − | <div class="column"> | + | <div class="column" style="float:right; text-align:center;"> |
| − | <img src="https://static.igem.org/mediawiki/2018/b/b0/T--SBS_SH_112144--Cyanobacteria13_.jpg" | + | <img src="https://static.igem.org/mediawiki/2018/b/b0/T--SBS_SH_112144--Cyanobacteria13_.jpg" style="width:500px; padding:20px 0px;"> |
| − | + | </div> | |
| + | </div> | ||
| − | < | + | <h4>Important Links!!!</h4> |
<a href='https://2018.igem.org/Team:SBS_SH_112144/Notebook '>Lab Notebook and Protocol</a > | <a href='https://2018.igem.org/Team:SBS_SH_112144/Notebook '>Lab Notebook and Protocol</a > | ||
Revision as of 16:59, 17 October 2018
Molecular cloning
1. We were able to use PCR to successfully amplify the synthesized gene (gel unshown)
2. Through homologous recombination using an exonuclease kit, we were able to fuse the cp-OS lysozyme 1 gene into the PET28A and PSB1C3 plasmid
3. We were able to cultivate the transformed BL21 E.coli and the addition of IPTG successfully triggered the expression of lysozyme.
Protein Purification
By purifying the protein using nickel column and later cleaving the His tag using TEV enzyme, we obtained pure lysozyme testified by the clearly labeled gel below.

Function test
1. By applying the lysozyme to a system(300ml in a microfuge tube reacting under different temperature) with bugbuster, cyanobacteria, pH buffer and water, we are able to observe cleavage effect illustrated by the OD660nm green light absorptance across large temperature and pH range. And through a series of mathematical computation and modeling, we are able to identify that the ideal pH is around 6.9 and temperature is at 37 °C.

2. After finding the ideal pH and temperature for the lysozyme, we successfully applied these elements to a new set of function test with different protein concentration and reaction time, and we are also able to, through PSO algorithm and 3D modeling in matlab, find out the optimal reaction time being 0.5h( quite different from the paper) and the protein concentration being 18 ug/mL.

3. Lastly, we built the prototype of our device to test the effectiveness of our device under the provided parameters to roughly mimic the actual commercial operation of such a device. And we were thrilled to see an over 70-80 percent of lysis even in the prototype.



