| Line 255: | Line 255: | ||
<p>The lactate dehydrogenase gene and the lactate transporter gene are combined in one circuit to achieve lactate production and transportation. For lactate dehydrogenase gene, we chose ldhD, and ldhDc is a codon-optimized version of ldhD, ldhDnARSdR is ldhD with D176A/I177R/F178S/N180R, and ldhDARSdR is the codon-optimized version of ldhDnARSdR. These codon optimizations are aim at increasing the production of lactate. The lldP protein gene is used to transport the lactate out of the cell. The lldP protein has 12 transmembrane alpha-helical segments and generally lack cleaved signal sequences and the lldP protein cotransports lactate with a proton[1]. At the same time, in order to detect the expression of these two genes, we add the Flag and 6 × His sequences respectively after the two.</p> | <p>The lactate dehydrogenase gene and the lactate transporter gene are combined in one circuit to achieve lactate production and transportation. For lactate dehydrogenase gene, we chose ldhD, and ldhDc is a codon-optimized version of ldhD, ldhDnARSdR is ldhD with D176A/I177R/F178S/N180R, and ldhDARSdR is the codon-optimized version of ldhDnARSdR. These codon optimizations are aim at increasing the production of lactate. The lldP protein gene is used to transport the lactate out of the cell. The lldP protein has 12 transmembrane alpha-helical segments and generally lack cleaved signal sequences and the lldP protein cotransports lactate with a proton[1]. At the same time, in order to detect the expression of these two genes, we add the Flag and 6 × His sequences respectively after the two.</p> | ||
| + | </div> | ||
| + | <div class="col-md-6 col-md-offset-3" style="text-align: center;"> | ||
| + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/b/be/T--HUST-China--2018-experimental_1-ldhD-lldP.png"> | ||
| + | <p>Figure 1. circuit of ldhD-lldP.</p> | ||
| + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/c/ce/T--HUST-China--2018-experimental_2-ldhDC-lldP.png"> | ||
| + | <p>Figure 2. circuit of ldhDC-lldP.</p> | ||
| + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/a/a9/T--HUST-China--2018-experimental_3-ldhDnARSdR-lldP.png"> | ||
| + | <p>Figure 3. circuit of ldhDnARSdR-lldP.</p> | ||
| + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/b/ba/T--HUST-China--2018-experimental_4-ldhDARSdR-lldP.png"> | ||
| + | <p>Figure 4. circuit of ldhDARSdR-lldP.</p> | ||
| + | |||
</div> | </div> | ||
| Line 262: | Line 273: | ||
</div> | </div> | ||
| − | + | <div class="col-md-6 col-md-offset-3" style="text-align: center;"> | |
| − | + | <img class="img-responsive" src="https://static.igem.org/mediawiki/2018/c/c4/T--HUST-China--2018-experimental_5-TH-gldA-lldP.png"> | |
| − | + | <p>Figure 5. circuit of TH-gldA-lldP.</p></div> | |
| − | + | ||
<div class="col-md-12"> | <div class="col-md-12"> | ||
Revision as of 20:31, 17 October 2018
Description
To convert optical energy into electric energy in a clean and sustainable way, Optopia is designed as a photovoltaic system consisting of photosynthetic microorganism (Rhodopseudomonas palustris) and electrogenic microorganism (Shewanella oneidensis). Synthetic biology strategies are applied to the system to trigger production and export of lactate in Rhodopseudomonas palustris, as well as to improve efficiency of lactate utilization and extracellular electron generation in Shewanella oneidensis. Compared to Cyanobacteria, also a kind of photosynthetic microorganism but generating oxygen in photosynthesis, Rhodopseudomonas palustris serves as a better carbon resource provider for Shewanella oneidensis, not only because of its anaerobic photosynthesis maintaining an anaerobic environment required for extracellular electron generation in Shewanella oneidensis, but also due to its capacity of reusing the waste from Shewanella oneidensis. Hence, functioning as a compatible and mutually beneficial optical MFC (Microbial Fuel Cell), Optopia creates a novel and optimized approach to utilize clean resources through optical-electric conversion.
Part1: Photosynthetic microorganism system

1.Cyanobacteria (Synechocystis)
Cyanobacteria are often chosen as engineering microorganism to convert optical energy into chemical energy. In our project, we chose Synechocystis PCC6803 for optical energy conversion. Cyanobacteria produce lactate for the electricity production of Shewanella. However, cyanobacteria itself lacks a pathway for producing lactate, and as a photoautotrophic microorganism, cyanobacteria lacks a lactate transporter to transport lactate out of the cell[1]. Therefore, we have designed two strategies to modify cyanobacteria.

Strategy 1:
The lactate dehydrogenase gene and the lactate transporter gene are combined in one circuit to achieve lactate production and transportation. For lactate dehydrogenase gene, we chose ldhD, and ldhDc is a codon-optimized version of ldhD, ldhDnARSdR is ldhD with D176A/I177R/F178S/N180R, and ldhDARSdR is the codon-optimized version of ldhDnARSdR. These codon optimizations are aim at increasing the production of lactate. The lldP protein gene is used to transport the lactate out of the cell. The lldP protein has 12 transmembrane alpha-helical segments and generally lack cleaved signal sequences and the lldP protein cotransports lactate with a proton[1]. At the same time, in order to detect the expression of these two genes, we add the Flag and 6 × His sequences respectively after the two.

Figure 1. circuit of ldhD-lldP.

Figure 2. circuit of ldhDC-lldP.

Figure 3. circuit of ldhDnARSdR-lldP.

Figure 4. circuit of ldhDARSdR-lldP.
Strategy 2:
In response to the preference for NADPH in the metabolism of cyanobacteria, transhydrogenase gene is used to achieve the goal of producing more NADPH, and the glycerol dehydrogenase gene is used to increase the lactate production. For transhydrogenase gene, we use TH gene, and gldA gene for glycerol dehydrogenase. The lldP protein gene is also used to transport the lactate out of the cell. And the same as strategy 1, the Flag and 6 × His sequences are used to detect the expression of the two genes.

Figure 5. circuit of TH-gldA-lldP.
After successfully constructing these gene circuits, the shuttle plasmid PCK306 is used to transform the genes into cyanobacteria, and the yellow fluorescent protein gene on the PCK306 plasmid is used to detect the transformation.
2.Rhodopseudomonas palustris
Rhodopseudomonas palustris is a kind of Purple Non-sulfur Bacteria. Under anaerobic conditions, Rhodopseudomonas palustris can utilize hydrogen, sodium thiosulfate, hydrogen sulfide as electron donors for photoautotrophic growth. Furthermore, it can also have heterotrophic growth under microaerobic to aerobic conditions [2]. In addition, Rhodopseudomonas palustris is widely used in sewage treatment, environmentally friendly, easily accessible [3]. Since it is capable of maintaining an anaerobic environment and reusing the acetate generated by Shewanella, Rhodopseudomonas palustris may be a better carbon source provider for Shewanella compared with cyanobacteria. Therefore, we decided to modify Rhodopseudomonas palustris so that it could produce lactate under the anaerobic condition and export it out of the cell. According to KEGG database [4], metabolic pathways, which are related to lactate metabolism in Rhodopseudomonas palustris, are as follows: .

To enhance the production of lactate in Rhodopseudomonas palustris, we plan to promote the conversion efficiency of pyruvate to D-lactate and malate to L-lactate. Therefore, we decide to transfer these two genes, mleS [5] and ldhA, [6] into Rhodopseudomonas palustris to generate more lactate intracelluarly. Considering the necessity of transporting lactate out of the cells, we also apply the gene lldP [7] encoding lactate permease. Since the order of these three genes in a polycistronic lead to different expression levels [8], we build a model to optimize the gene order to maximize the gene expression. Also, we use codon optimized tool Jcat [9] to optimize the codons of these genes.

mleS:malate dehydrogenase, the conversion of malic acid to L-lactate.
ldhA:fermentative D-lactate dehydrogenase, NAD-dependent, convert pyruvate to D-lactate.
lldP:L-lactate permease, the lactate is transported out of the cell.
To validate our modeling results, we continue to build three different gene circuits which can determine the highest lactate production efficiency of our total circuit:



Part2: Electrogenic microorganism system

1. Shewanella oneidensis
Shewanella oneidensis is becoming more and more popular these years as it could transfer electron to the electrode and generate electricity. This fascinating power mainly attributes to a number of conductive c-type cytochromes (c-Cyts) including OmcA-MtrCAB and CymA. CymA could conduct electrons to the MtrCAB complex. Then, the bacteria would use its extension or vesicles of the outer membrane and periplasm to transfer outer membrane c-type cytochromes.

The amount of electricity produced by Shewanella is closely related to the bacteria’s metabolism.
①. Glycolysis: Glycolysis is the metabolic pathway that converts glucose C6H12O6 into pyruvate. As glyceraldehyde-3-phosphate turns into 1,3-Bisphosphoglycerate, NADH is generated, which would be used to transfer electron.
②. TCA cycle: TCA cycle is a series of chemical reactions happened in mitochondrion. The reactions of the cycle are carried out by eight enzymes and three of them including malate dehydrogenase could help to produce NADH.
③. Pyruvate fermentation: Pyruvate fermentation is a common metabolic pathway in bacteria. Several steps of pyruvate fermentation could also produce NADH and the related enzymes are pyruvate formate-lyase, lactate dehydrogenase and formate dehydrogenase.

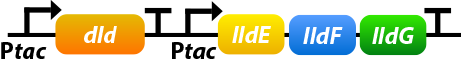
Shewanella oneidensis MR-1 prefers to use lactate as its carbon source since the amount of lactate-based biomass is more than acetate-based biomass or pyruvate-based biomass. Dld and lldEFG are D- and L-lactate dehydrogenase enzymes, which is the first step of utilizing lactate. To make the use of lactate more efficiently, we overexpress four genes: dld, lldE, lldF, lldG.[10]
①. dld: dld refers to FAD-dependent D-lactate dehydrogenase which could catalyze D-lactate’s transformation into pyruvate.
②. lldEFG: They could encode a L-lactate dehydrogenase complex which could catalyze D-lactate’s transformation into pyruvate.
To ensure that the genes would be expressed efficiently, we add a promoter before lldEFG:
NADH is a significant part of extracellular electron transfer(EET) as it could carry electron. Strenghthening the regeneration of NADH would make EET more efficiently.
To achieve this goal, we overexpress these four genes: gapA2, mdh, pflB, fdh. [11]
①. gapA: It encodes glyceraldehyde-3-phosphate dehydrogenase which could transform 3- phosphoglyceraldehyde into 1,3- diphosphoglycerate.
②. mdh: It encodes NAD dependent malate dehydrogenase which transforms malate into pyruvate
③. pflB: It encodes pyruvate formate-lyase to transform pyruvate into Acetyl-CoA.
④. fdh: It encodes formate dehydrogenase to transform formate into CO2..
Also, to ensure that the genes would be expressed efficiently, we add an promoter before pflB and fdh:

Part3: Whole design

Design of MFC
We have designed a bipolar chamber MFC this year. Proton exchange membrane divided it into anode chamber and cathode chamber. Anode chamber containing S.oneidensis, nutrient substance(LB、lactate ) or other electrical producing microbes were sealed to prevent the entry of external oxygen. Considering safety and oxidation-reduction potential, we put ferric chloride solution in cathode chamber so that S.oneidensis can transfer electrons outside of their membranes by electron transport chain. Then electrons will reduce ferric ion into ferrous through carbon cloth and produce electricity. We recorded open circuit voltage curve and load voltage curve of MFCs in each different systems. Also, we have measured the biomass of each system in order to ensure whether the improved electricity could be attributed to more attached Shewanella cells on the anodes or the higher electroactivity of single cell.[11]

Co-culture
Obviously, the ecological relationship between microorganisms is very complex. There is not only the competition between them for the nutrient, but also the regulation of metabolites among them including induction, transgenosis and synergistic metabolism. Besides, it has been found that the co-culture of microorganisms can improve the electric efficiency of Microbial Fuel Cell under certain conditions.
Metabolites exchange is a common relationship in co-culturing. Therefore, we have designed a clear microbial metabolic pathway to achieve the conversion from light to electricity as well as used more potential symbiotic relationships between the flora to help improve the electricity production efficiency of MFC.
By consulting literature, we found two kinds of microorganisms——Cyanobacteria and Rhodopseudomonas palustris, both of which can utilize light energy and provide lactate to S.oneidensis after doing molecular construction.
In order to provide a basic growth environment, we mix the culture medium of different strains.(Please refer to our protocol section for the composition of the mediums.)
Synechocystis PCC6803
Lactate produced by Synechocystis PCC6803 can be used as the optimal carbon source for Shewanella. At the same time, acetate produced by Shewanella can be used as the organic carbon source of Synechocystis PCC6803 to increase the lactate production. And the metabolite exchange of Synechocystis PCC6803 and Shewanella is the basis for our photoautotrophic MFC.[12].
Rhodopseudomonas palustris
We attempted to engineer Rhodopseudomonas palustris by synthetic biology to achieve the same or a better function of Synechococcus elongatus.
In the preliminary experiment, we found that there may be more potential interactions in the co-culture of Rhodopseudomonas palustris and Shewanella, which can greatly improve the coulombic efficiency of our MFC (please refer our results section for more detials). This is an unexpected surprise for us, which improve to our confidence in the success of the project.
Reference
[1]Henrike Niederholtmeyer, Bernd T. Wolfstädter, David F. Savage, Pamela A. Silver, Jeffrey C. Way. Engineering Cyanobacteria to Synthesize and Export Hydrophilic Products. Applied and Environmental Microbiology. 2010, 76(11): 3462-3466.
[2] Research progress and application of Rhodopseudomonas palustris [Journal] Wang Yuexian, Liu Dehai - Henan Animal Husbandry and Veterinary Medicine (Comprehensive Edition) 2015, Issue 1
[3] ZHOU Maohong, ZHAO Xiaowei, WU Xuechang. Study on the ability of photosynthetic bacteria Rhodopseudomonas palustris to assimilate phosphorus[J]. Bulletin of Science and Technology, 2002(02): 142-146.
[4] KEGG, https://www.genome.jp/kegg/pathway.html
[5] Cloning and sequence analysis of the gene encoding Lactococcus lactis malolactic enzyme: relationships with malic enzymes FEMS Microbiol Lett. 1994 Feb 1;116(1):79-86
[6] Fine tuning the transcription of ldhA for d-lactate production August 2012, Volume 39, Issue 8, pp 1209–1217
[7] Transport of L-Lactate, D-Lactate, and Glycolate by the LldP and GlcA Membrane Carriers of Escherichia coli Volume 290, Issue 2, 18 January 2002, Pages 824-829
[8] Enhancement of Hydrogen Production and Carbon Fixation in Purple Nonsulfur Bacterium Bacterium by Synthetic Biology Shou-Chen Lo
[9] Jcat, http://www.jcat.de/#opennewwindow
[10] Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization PNAS February 24, 2009 106 (8) 2874-2879
[11] Modular Engineering Intracellular NADH Regeneration Boosts Extracellular Electron Transfer of Shewanella oneidensis MR‑1 ACS Synth. Biol. 2018, 7, 885−895
[12]Varman A. M., Yu Y., You L. & Tang Y. J. Photoautotrophic production of d-lactate in an engineered cyanobacterium. Microb. Cell Fact. 12, 117 (2013).



