Our Project
Overview
Shewanella oneidensis is an interesting species of bacteria that not only can survive in anaerobic conditions but can also generate energy. These characteristics makes it an impressive microbe to use in microbial fuel cells. In addition, it feeds off of lactate which can then increase the growth of Shewanella. By genetically engineering E.coli to synthesize lactate, we can determine if there is a symbiotic relationship between these types of bacteria.
Background
Greenhouse gases produced from the burning of coal and other fossil fuels are damaging our atmosphere. In the rivers of Pittsburgh are microorganisms that can be used to create microbial fuel cells (MFCs). In MFCs the microorganisms transfer electrons to reduce metals and if a conducting wire is connected to the metal cathode, current will run through the wire and the electricity can be utilized. The goal was to create energy on demand by engineering E. coli to produce a carbon source for an electrogenic bacteria. Shewanella oneidensis (Shewanella species isolated from Lake Oneida, New York) is a laboratory model strain of electrogenic bacteria and he complete genome of Shewanella is available. The preferred carbon source of Shewanella is lactate (Brutinel and Grainick, 2012) and the enzyme that is involved in lactate synthesis in Shewanella is the ldha gene (Le Laz et al. 2016).
The Evolution of the Microbial Fuel Cell
A microbial fuel cell, MFC, utilizes chemical energy to produce electricity from catalytic reactions from microorganisms. Bacteria are able to oxidize aerobic matter and generate electricity. We can then use that electricity to power many things such as lemon clocks and LED lights. Prior to joining the iGEM team, some of our team members participated in the Ice-T program at The Citizen Science Lab and learned to use coding language to analyze the biochemistry and physiology of a microbial fuel cell made of mud. We have now evolved from the MFC made out of mud into an MFC that is created out of media, and may be the site of a possible symbiotic relationship. We have also improved on the anode used in our MFC. During Ice-T, we would use a piece of zinc that we would tie to our positive wire and the zinc would be our anode. With the help of Bharat Gattu, a PhD student at the University of Pittsburgh, we were able to create a copper foam to use as our anode. The idea behind the foam is that provides an increase in surface area which allows for more space for the Shewanella biofilm. In future experiments, we would like to test how effective the copper foam is in increasing the surface area for Shewanella biofilm growth.
Methods
Shewanella oneidensis MR-1 was obtained from ATCC (spec sheet shown below).


The Shewanella ldhA gene sequence was identified. Below is the Genbank information.

A gBLOCK was ordered from IDT to express ldhA constitutively, the sequence is shown below. (BBa_K2625001)

The gBLOCK was PCR amplified.
The gBLOCK and backbone pSB1C3 were digested with EcoRI/PstI and gel purified.
Colonies were screened by colony PCR (lane 1, molecular weight markers, lane 2-9 clones, lane 10 vector).

The insert was confirmed by sequencing.
We wanted to grown E. coli with the plasmid and Shewanella together so Shewanella needed to be transformed however multiple attempts to transform Shewanella to chloramphenicol resistance were unsuccessful (TSS, liquid nitrogen protocol (Tripp et al. 2013)).
Microbial Fuel Cell Setup
MM3 media recipe (Le Laz et al. 2016).
Shewanella without lactate, with lactate, with E. coli + ldhA

New Part Shewanella ldhA gene BBa_K2565001
This composite part is used to constitutively express high levels of Shewanella oneidensis MR-1 fermentative lactate dehydrogenase NADH dependent ldhA (BBa_K2565000) in E. coli, locus tag SO_0968.

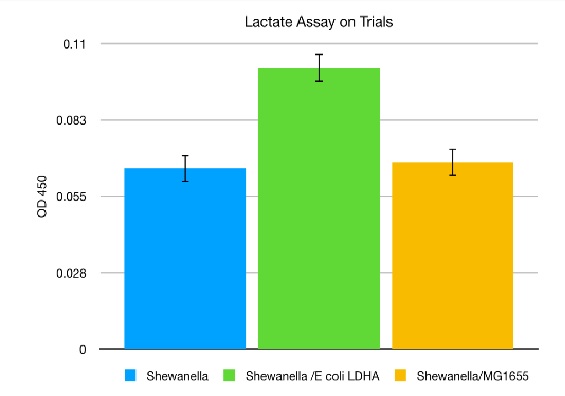
The left figure shows that the media with E. coli constitutively expressing ldhA contains more lactate than LB only. The right figure shows the amount of lactate in the media of cultures so that when Shewanella and E. coli are grown together there is more lactate in the media. The MG1655 strain of E. coli does not express the Shewanella ldhA gene.

This graph shows the voltage produced by microbial fuel cells (MFC) in 15 hours. The MFC with Shewanella and E. coli produced more voltage than Shewanella alone. The MFC with Shewanella and E. coli expressing Shewanella ldhA produced more consistent voltage than E. coli without Shewanella ldhA.
References
Brutinel ED, Gralnick JA. Preferential utilization of D-lactate by Shewanella oneidensis. 2012.Appl Environ Microbiol. 2012 Dec;78(23):8474-6. doi: 10.1128/AEM.02183-12.
Le Laz S, Kpebe A, Bauzan M, Lignon S, Rousset M, Brugna M. 2016. Expression of terminal oxidases under nutrient-starved conditions in Shewanella oneidensis: detection of the A-type cytochrome c oxidase.Sci Rep. 2016 Jan 27;6:19726. doi: 10.1038/srep19726.
Tripp VT, Maza JC, Young DD. 2013. Development of rapid microwave-mediated and low-temperature bacterial transformations.J Chem Biol. 2013 May 21;6(3):135-40. doi: 10.1007/s12154-013-0095-4.
