| Line 59: | Line 59: | ||
</section> <!-- end s-home --> | </section> <!-- end s-home --> | ||
| − | <section id=' | + | <section id='Intro' class="s-services"> |
<div class="row section-header has-bottom-sep" data-aos="fade-up"> | <div class="row section-header has-bottom-sep" data-aos="fade-up"> | ||
| Line 304: | Line 304: | ||
<img src="https://static.igem.org/mediawiki/2018/c/c6/T--Newcastle--MeasurementFigure5.jpg"style="width:30%"> | <img src="https://static.igem.org/mediawiki/2018/c/c6/T--Newcastle--MeasurementFigure5.jpg"style="width:30%"> | ||
| − | <p> <center><b>Figure 5 | + | <p> <center><b>Figure 5. Impact of downscaling competent cell production from 50 mL falcon tube to 400 uL 96 plate well volumes. Black circles = 50 mL falcon tube, grey squares = 2 mL microcentrifuge tubes, light grey triangle = 400 uL 96 well plate.</b> Competent cells were produced using MgCl2+CaCl2 protocol and transformed using standard heat shock. 100 uL of transformed cells were then plated out on SOB+CAM and incubated overnight at 37 degrees. Colonies were counted and transformation efficiency (TrE) calculated. A significant difference in transformation efficiency depending on reaction vessel was shown (ANOVA, F2,15 = 8.24, P = 0.004). Post hoc Tukey test indicated that both 50 mL Falcon tubes and 2 mL microcentrifuge tube volumes had a statistically insignificant difference in TrE (T = 0.06, p = 0.998). 96 well plate TrE was statistically lower than both 50 mL and 2 mL volumes (p = 0.009 and p = 0.008 respectively). Plasmid concentration had a significant effect on TrE, with the 100 pg/uL TrE a power of 10 greater on average (t= -2.81, d.f = 16, p = 0.013). </p> |
<p> To reduce protocol complexity and length, wash number and wash combination were evaluated, with the results suggesting no significant impact on transformation efficiency (Figure 6). Wash steps were excluded moving forward to streamline and decrease protocol complexity, without significant loss of transformation efficiency. </p> | <p> To reduce protocol complexity and length, wash number and wash combination were evaluated, with the results suggesting no significant impact on transformation efficiency (Figure 6). Wash steps were excluded moving forward to streamline and decrease protocol complexity, without significant loss of transformation efficiency. </p> | ||
| Line 310: | Line 310: | ||
<img src="https://static.igem.org/mediawiki/2018/9/96/T--Newcastle--MeasurementFigure6.jpg"style="width:30%"> | <img src="https://static.igem.org/mediawiki/2018/9/96/T--Newcastle--MeasurementFigure6.jpg"style="width:30%"> | ||
| − | <p> <center><b>Figure 6 | + | <p> <center><b>Figure 6. Effect of different wash steps on overall transformation efficiency (TrE). All competent cell preparation followed the standard MgCl2-CaCl2 protocol, with only the wash steps altered.</b> 0 Wash - initial culture pellet followed by immediate aliquot of 100 uL storage/transformation buffer. 1 wash – a combined 100 mM MgCl2 and 100 mM CaCl2 buffer with 1 wash step. MgCl2 + CaCl2 – a combined 100 mM MgCl2 and 100 mM CaCl2 buffer with original two wash steps. MgCl2/CaCl2 – a 100 mM MgCl2 wash step, followed by a separate 100 mM CaCl2 wash step as per standard protocol. No significant impact on TrE (Kruskal-Wallis, H = 1.34, d.f. = 3, p = 0.720) was shown. Removing the wash step was the most effective (mean TrE = 2.30 x 106), with the more time consuming MgCl2-CaCl2 protocol being the second most effective (mean = 2.18 x 106). The least effective were the combined MgCl2/CaCl2 two wash (mean TrE = 1.79 x 106) and one wash (mean TrE = 1.66 x 106).</p> |
| Line 356: | Line 356: | ||
<img src="https://static.igem.org/mediawiki/2018/3/37/T--Newcastle--MeasurementFigure7.jpg"style="width:30%"> | <img src="https://static.igem.org/mediawiki/2018/3/37/T--Newcastle--MeasurementFigure7.jpg"style="width:30%"> | ||
| − | <p> <center> <b>Figure 7 | + | <p> <center> <b>Figure 7. Initial DoE scoping test of low, medium and high transformation buffer concentrations with different cryoprotectants.</b> Black – 7.5% DMSO, grey – 18% glycerol. Low Buffer concentration consisted of 15 mM CaCl2.2H2O solution. High buffer concentration consisted of a 100 mM MgCl2.6H2O, 100 mM CaCl2.6H2O. 10 mM kOAc, 100 mM MnCl2.4H2O, 100 mM RbCl, 100 mM NiCl2, 3 mM [Co(NH3)6]Cl3 and 100 mM KCl solution. Two-way ANOVA determined a significant difference in TrE dependent on Buffer Concentration (ANOVA, Buffer Concentration: F2,16 = 4.593, p = 0.0265) whilst there was no significant difference between cryoprotectants (ANOVA, Cryoprotectant: F1,16 = 3.469, p = 0.0810). There was a significant interaction between the two (ANOVA: Interaction: F2,16 = 6.548, p = 0.0084). Post hoc Tukey test confirmed that the medium wash concentration with DMSO resulted in significantly greater TrE (mean TrE = 9.29 x 106) whilst all other TB compositions were insignificantly different.</p></center> |
| Line 385: | Line 385: | ||
<img src="https://static.igem.org/mediawiki/2018/2/23/T--Newcastle--MeasurementFigure8.jpg"style="width:30%"> | <img src="https://static.igem.org/mediawiki/2018/2/23/T--Newcastle--MeasurementFigure8.jpg"style="width:30%"> | ||
| − | <p> <center><b>Figure 8 | + | <p> <center><b>Figure 8. Assessment of pH buffer effect on overall transformation efficiency (TrE). </b> Transformation buffer used was the medium scoping buffer (MSB) with 7.5% DMSO. Control – MSB without pH buffer, HEPES – MSB + 10 mM HEPES, PIPES – MSB + 10 mM PIPES, MES – MSB + 10 mM MES, MOPS – MSB + 10 mM MOPS. All buffers were adjusted to 6.8 pH for comparison and to prevent manganese dioxide from precipitating out of the MSB. Inclusion of pH buffering agent significantly affected TrE (ANOVA, F4,10 = 6.45, p = 0.008) (Figure 9). Post hoc Tukey test clarified that HEPES, PIPES and MES had a significant increase in TrE when compared to the control (p = 0.006, p = 0.032, p = 0.035 respectively). MOPS had minimal effect on TrE when compared with the control with no significant difference being shown (p = 0.234), yet mean TrE was still 4.80 x 105 greater than control.</p></center> |
</div> | </div> | ||
| Line 430: | Line 430: | ||
| − | <p><center><b>Figure 9 | + | <p><center><b>Figure 9. Comparison between initial unoptimised and further optimised automated competent cell and transformation protocols.</b> Circles indicate raw data values, with central line indicating mean with SD error bars. Both protocols followed the same 0 wash method. A significant increase in TrE was shown, with a mean TrE of 1.20 x 105 (Mann-Whitney, U = 333.0, n. = 21,30, p < 0.001).</p> |
| Line 475: | Line 475: | ||
<img src="https://static.igem.org/mediawiki/2018/5/59/T--Newcastle--MeasurementFigure10.jpg"style="width:60%"> | <img src="https://static.igem.org/mediawiki/2018/5/59/T--Newcastle--MeasurementFigure10.jpg"style="width:60%"> | ||
| − | <p><center><b>Figure 10 | + | <p><center><b>Figure 10. Optimised workflow for the automated transformation buffer optimisation and transformation efficiency (TrE) analysis protocol (ATBOT).</b> Light grey boxes indicate manual steps whereas dark grey boxes indicate automated steps. Y/N show a logic step in python script. All liquid handling was carried out by the OT-2 robot, with TempDeck module allowing for temperature control and heatshock steps without manual interaction. Box (*) describes the logic steps that are undertaken during the for and if/else/elif loops required for the OT-2 to carry out complex P10/P300 pipetting steps. Outcomes of ATBOT are highlighted as 1, 2 or 3. Outcome 1 allows for the assessment of accurate CFU and calculation of TrE which can be inputted into JMP Pro and used to further model the DoE design space. Outcome 2 allows for successfully transformed colonies to be isolated for further testing or use. Outcome 3 transfers post-recovery transformants into a selection broth, allowing for either overnight incubation or plate reader assessment. Plate reader assessment can be used to determine growth rates and may potentially be used as a means to accurately calculate TrE (********section 1.8.1**********WHAT IS THIS?).</p> |
</div> | </div> | ||
| Line 559: | Line 559: | ||
<img src="https://static.igem.org/mediawiki/2018/a/a4/T--Newcastle--MeasurementFigure11.jpg"style="width:60%"> | <img src="https://static.igem.org/mediawiki/2018/a/a4/T--Newcastle--MeasurementFigure11.jpg"style="width:60%"> | ||
| − | <p><center><b>Figure 11 | + | <p><center><b>Figure 11. Prediction profiler modelling the effect that varying reagent concentration has on overall transformation efficiency (TrE). </b> Overall prediction profile set to most desirable transformation buffer composition. Black line indicates concentration relative to predictive TrE. Vertical dashed red line indicates the concentration of reagent at most desirable composition while horizontal dashed red line indicates predicted average TrE at desirable composition. Top panels show calculated effects of reagents after TrE was calculated after a 37℃ overnight 16 hour incubation post transformation recovery step. Bottom panels show calculated effects of reagents after TrE was recalculated after 96 hour incubation at room temperature (22-25℃) post transformation recovery step. Highlighted green panel indicate that a higher MgCl2.6H2O concentration had a statistically significant (p=0.0096) positive increase in overall TrE after a 96 hour incubation. Its effect after 16 hours incubation was shown to be positive, however not statistically significant. Highlighted blue panels indicate a statistically significant decrease in overall TrE when concentration is increased. Blue panel (A) indicates that the presence NiCl2 is significantly inhibitory to overall TrE (p = 0.0058) after 16 hours post recovery. Blue panel (B) suggests that CaCl2.6H2O has a negative impact with increasing concentration (p = 0.0490) and blue panel (C) indicates that kOAc has a significant inhibitory effect (p = 0.0153) on overall TrE after 96 hours post incubation. [Co(NH3)6]Cl3 at both post transformation time points is shown to have negligible effect on TrE. </p></center> |
<img src="https://static.igem.org/mediawiki/2018/e/ec/T--Newcastle--MeasurementFigure12.jpg"style="width:50%"> | <img src="https://static.igem.org/mediawiki/2018/e/ec/T--Newcastle--MeasurementFigure12.jpg"style="width:50%"> | ||
| − | <p><center><b>Figure 12 | + | <p><center><b>Figure 12. Prediction profile modelling the interactions of transformation buffer (TB) constituents deemed to significantly affect overall transformation efficiency (TrE) after 16 hours of post-recovery incubation. </b>Reagents were selected based on their significance, with only reagents with individual P values < 0.1 being selected for modelling. (A) Prediction profile set to most desirable TB composition. Black line indicates concentration relative to predictive TrE. Vertical dashed red line indicates the concentration of reagent at most desirable composition while horizontal dashed red line indicates predicted average TrE at desirable composition. Highlighted blue panels indicate a significant inhibitory effect on TrE. Highlighted green panels indicate significantly positive increase in TrE. Both MgCl2.6H2O and RbCl have a positive interaction with increasing concentration (P = 0.0095). NiCl2 and DMSO have a significant inhibitory interaction with individual increasing concentration (P = 0.0002 and P = 0.0479), however when both concentrations are increased they have a positive interaction with each other (P = 0.0054). (B) Interaction profile describing notable interactions between all significant reagents. Each panel represents an interaction between two reagents, with one reagent set at maximum concentration and the other at either lowest concentration (red line) or highest concentration (blue line). Panels positioned above or below the reagents in the graph are the reagents at maximum concentration, whereas panels positioned left or right are the reagents at either low or high concentrations. All other reagents included in the model but are not being examined in the panel are set to maximum. </p></center> |
</div> | </div> | ||
| Line 600: | Line 600: | ||
<img src="https://static.igem.org/mediawiki/2018/2/2c/T--Newcastle--MeasurementFigure13.jpg"style="width:70%"> | <img src="https://static.igem.org/mediawiki/2018/2/2c/T--Newcastle--MeasurementFigure13.jpg"style="width:70%"> | ||
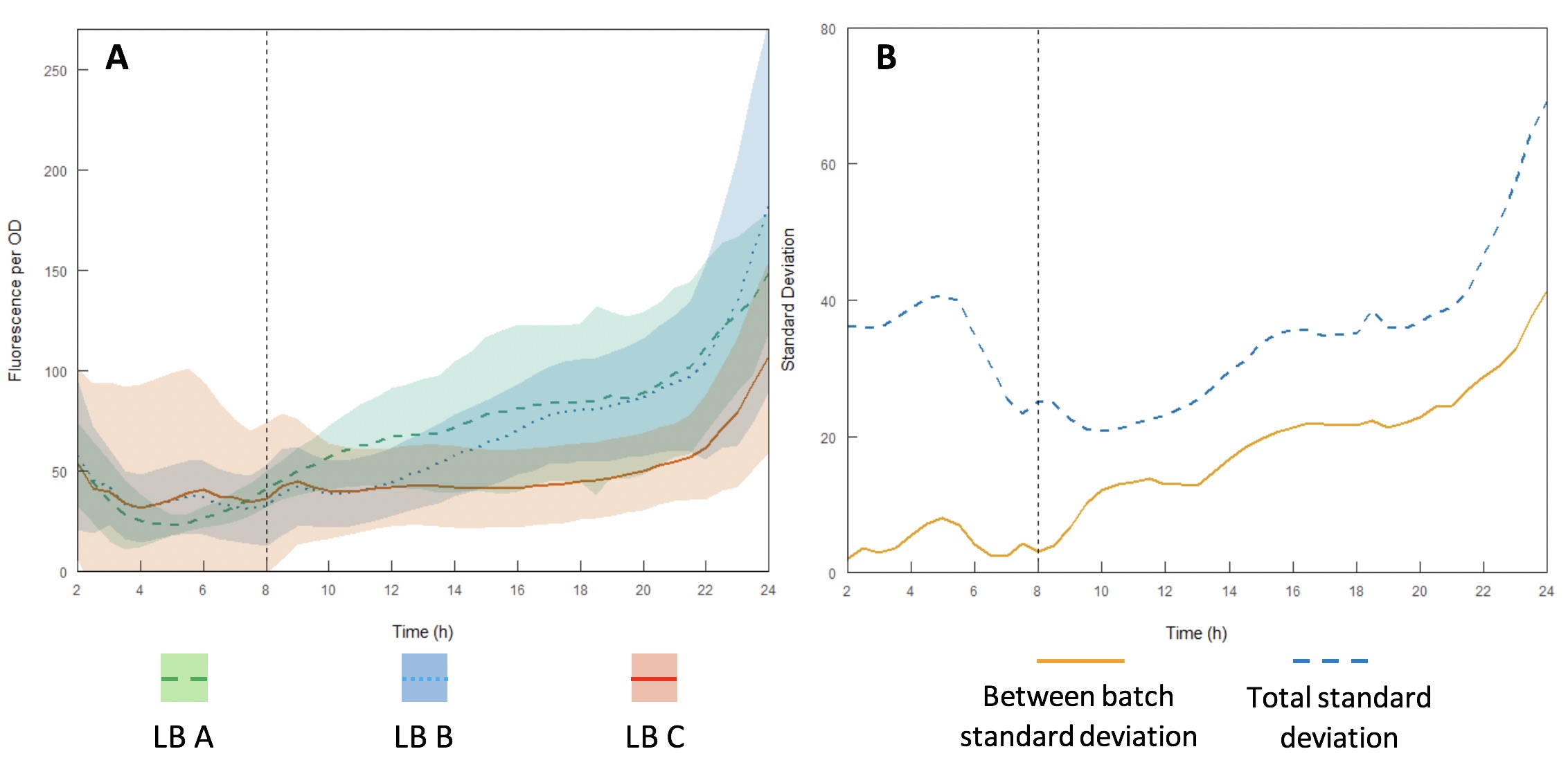
| − | <p><center><b>Figure 13 | + | <p><center><b>Figure 13. A) fluorescence per OD600 for recombinant <i>E. coli</i> DH5α </b> expressing the iGEM interlab positive control device GFP gene E0040 in three batches (designated A, B and C) of LB broth made with tryptone and yeast extracts from different batches and different manufacturers with standard deviations (shaded areas); B: standard deviation of fluorescence per OD between each batch and total standard deviation across the whole dataset. Dotted vertical line represents the end of exponential growth phase and start of stationary phase.</p> </center> |
</div> | </div> | ||
| Line 624: | Line 624: | ||
<img src="https://static.igem.org/mediawiki/2018/a/aa/T--Newcastle--MeasurementFigure14.jpg"style="width:50%"> | <img src="https://static.igem.org/mediawiki/2018/a/aa/T--Newcastle--MeasurementFigure14.jpg"style="width:50%"> | ||
| − | <p><center><b>Figure 14 | + | <p><center><b>Figure 14. Growth curves for <i>E. coli</i> DH5α in rich defined media DoE runs over 22 hours.</b></p></center> |
<p>The OD600 values at 24 hours were used to build a Projection to Latent Structures (or Partial Least Squares (PLS)) model using JMP software. This allowed visualisation of factor effects on E. coli growth (Figure 15). It is observed that there are positive effects on growth of increasing citrate, thiamine and tricine concentration, while there are negative effects of increasing K2HPO4. Additionally, four interactions between components in the media were shown to be important predictors of DH5α growth. These are interactions between citrate and tricine (VIP = 1.45), thiamine and EDTA (VIP = 1.27), thiamine and tricine (VIP = 1.20) and citrate and thiamine (VIP = 1.11). The media composition predicted by the model to maximise DH5α growth is shown in Figure 15.</p> | <p>The OD600 values at 24 hours were used to build a Projection to Latent Structures (or Partial Least Squares (PLS)) model using JMP software. This allowed visualisation of factor effects on E. coli growth (Figure 15). It is observed that there are positive effects on growth of increasing citrate, thiamine and tricine concentration, while there are negative effects of increasing K2HPO4. Additionally, four interactions between components in the media were shown to be important predictors of DH5α growth. These are interactions between citrate and tricine (VIP = 1.45), thiamine and EDTA (VIP = 1.27), thiamine and tricine (VIP = 1.20) and citrate and thiamine (VIP = 1.11). The media composition predicted by the model to maximise DH5α growth is shown in Figure 15.</p> | ||
| Line 630: | Line 630: | ||
<img src="https://static.igem.org/mediawiki/2018/d/d4/T--Newcastle--MeasurementFigure15.jpg"style="width:70%"> | <img src="https://static.igem.org/mediawiki/2018/d/d4/T--Newcastle--MeasurementFigure15.jpg"style="width:70%"> | ||
| − | <p><center><b>Figure 15 | + | <p><center><b>Figure 15. Prediction profiler for the model of factors affecting growth of <i>E. coli</i> DH5α in a defined rich medium after 24 hours.</b> The vertical red dashed line represents the factor concentration or level in the medium, changing which alters the horizontal red dashed line, representing culture OD600. Statistically significant factors highlighted in green were citrate, thiamine, tricine and KH2HPO4 (VIP = 2.39, VIP = 2.14 and VIP = 1.99, VIP = 1.04 respectively). Factor levels are set to max desirability, shown in red text beneath each x-axis (i.e. levels producing the maximum OD600). The model was constructed using JMP Pro v.12 statistical software (SAS Institute Inc.), based on experimental results of a definitive screening based experimental design.</p></center> |
<p>From the results here, and from the previously shown membrane permeability altering characteristics of EDTA, it is likely that interactions would be observed with other media components which require active transport into the cell. As such, for the further development of a rich defined media for recombinant expression in DH5α, and with the aim of reducing variability, it would be recommended that EDTA and citrate be omitted - the model produced here predicts good growth is possible without these components. Tricine on the other hand appears to be an excellent buffer for resisting the decreasing media pH as a result of metabolism and inclusion of which is recommended in further development of this media.</p> | <p>From the results here, and from the previously shown membrane permeability altering characteristics of EDTA, it is likely that interactions would be observed with other media components which require active transport into the cell. As such, for the further development of a rich defined media for recombinant expression in DH5α, and with the aim of reducing variability, it would be recommended that EDTA and citrate be omitted - the model produced here predicts good growth is possible without these components. Tricine on the other hand appears to be an excellent buffer for resisting the decreasing media pH as a result of metabolism and inclusion of which is recommended in further development of this media.</p> | ||
Revision as of 12:12, 17 October 2018