| Line 193: | Line 193: | ||
</div> | </div> | ||
<div class="tab-pane fade" id="Demo_Part4" role="tabpanel"> | <div class="tab-pane fade" id="Demo_Part4" role="tabpanel"> | ||
| − | <h2 class="heavy"> | + | <h2 class="heavy">A workflow to produce functionalized cellulose</h3> |
<hr/> | <hr/> | ||
| − | <p> | + | <p>This project was not only about functionalizing cellulose but ambitioned to define the whole process from component purification to production <i>in vivo</i> (Figure 1).</p> |
| − | + | <div style="text-align:center"> | |
| + | <figure id="figure14" class="figure" style="text-align:center;"> | ||
| + | <img style="width : 80%; heigth = auto;" src="https://static.igem.org/mediawiki/2018/7/72/T--Toulouse-INSA-UPS--Demonstrate--Youn--WorkFlow.png" class="figure-img img-fluid rounded" alt="Fluorescence Retained"> | ||
| + | <figcaption id="figcaption14" class="figure-caption"><strong>Figure 1: </strong><i> Workflow of our Cerberus project experimental validation process. </i> | ||
| + | </figcaption> | ||
| + | </figure> | ||
</div> | </div> | ||
| − | </ | + | <p>Indeed, we have defined and validated the basis to model our platform. This allows testing in silico any further modifications that could be of interest for Cerberus. Next, we demonstrated the efficiency of the Cerberus production in <i>E. coli</i>. This validated our choices for the strain, vector and culture conditions (IPTG, AzF, arabinose…). Then, we also successfully purified Cerberus and shown this through SDS-PAGE and Western Blot. As previously described, production of biotinylated compounds was validated as well as cellulose functionalization using Cerberus. We went even further and started the production of cellulose <i>in vivo</i> with <i>Gluconacetobacter hansenii</i>. The next step would have been to demonstrate the possibility to co-culture this strain and a strain synthesizing Cerberus and the function. Then, the final stage will be to start the production of cellulose <i>in vivo</i>. Even if we did not have enough time to each this final step, it was thought from the beginning of the project, as it was for this whole workflow.</p> |
<!--CONTENT ENDS HERE--> | <!--CONTENT ENDS HERE--> | ||
Revision as of 19:35, 16 October 2018
DEMONSTRATE
Cerberus was a huge challenge on many levels. From the modelling to the in vitro validations, from functionalizing cellulose to laying the ground for in vivo production, we went a lot farther than we had dreamed about. Both functionalities of the Cerberus platform and its applicability have been demonstrated. This page summarizes these achievements.

Validation of the Three Binding Heads
Cerberus is composed of three different domains: CBM3a for cellulose binding, mSA2 for biotin affinity and AzF to click molecules with an alkyne moiety. They all have been demonstrated sequentially.
 The CBM3a head and cellulose binding
The CBM3a head and cellulose binding
The cellulose binding property was based on the CBM3a cellulose-binding domain. To demonstrate its functionality, we fused it to the mRFP1 fluorescent protein. Our results clearly proved its efficiency to bind cellulose (Figure 1). This was the first step from which we built the Cerberus platform.

 The mSA2 domain and biotin binding
The mSA2 domain and biotin binding
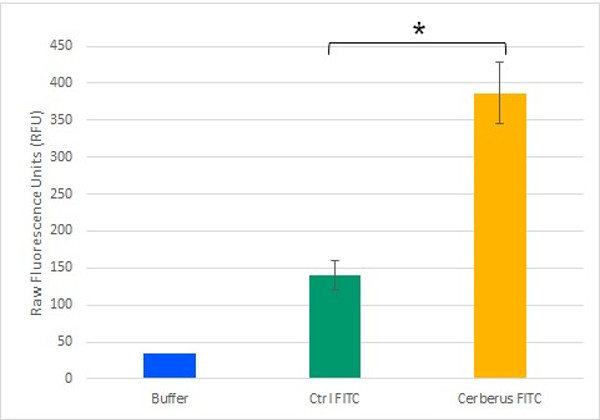
To demonstrate the streptavidin head functionality, we combined it to the already demonstrated CBM3a domain. We then validated this construction with biotinylated fluorophores from both in vivo (Figure 2) or in vitro origins. This also demonstrated the capacity of the construction to add new properties to cellulose.

 The AzF head and click chemistry
The AzF head and click chemistry
To assess the functionality of the AzF head, we used the previous construction with CBM3a and streptavidin and added the UnAA to the second linker (i.e, the whole Cerberus platform). We confirmed the activity of this integration using DBCO-Fluorescein as showed in Figure 3, but also DBCO-paramagnetic beads. This completed the validation of our system.
Adding new properties to cellulose
 Color
Color
It was not initially planned to add colors to cellulose. However, mRFP1 has the property to provide a red hue without excitation. This was easily observable on bacterial cellulose after incubation with CBM3a-mRFP1 (Figure 1). This could be of special interest since a lot of proteic dyes are available (see iGEM Imperial 2016). The current dyes used in the textile industry are often toxic to people or the environment. Cerberus association with proteic dyes could thus be a safer alternative solution.


 Fluorescence
Fluorescence
The first sought-after property of mRFP1 is for sure fluorescence. We successfuly produced fluorescent cellulose by combining cellulose, our platform and fluorescent dyes in several ways : by adding biotinylated fluorescent compounds, by clicking fluorescent molecules with an alkyne function or even by testing the CBM3a-mRFP1 construction on bacterial cellulose (Figure 2). This proves that physico-chemical properties of protein could be transfered to cellulose and allow to imagine many other developments (luminescence for instance).


 Antibiotic
Antibiotic
We also conjugated an antimicrobial peptide, scygonadin, to the streptavidin. The first results seemed to indicate this could work (Figure 3) but it was not as a definitive result as the other functionalization assays we have performed. This will require further testings since such a functionalization arouse very important expectations from the public (see Human Practices).

 Paramagnetism
Paramagnetism
Last but not least, we validated the grafting of paramagnetic beads to cellulose (Figure 4). So, our Cerberus platform has allowed us to create paramagnetic cellulose. In our opinion, this is the functionalization that should permit the largest panel of applications, especially in combination with other functions as Cerberus allows. Just imagine all the application in purification of compounds or cells!

Perspectives and conclusion
We successfully set up a workflow allowing the synthesis of Cerberus, a proteic platform that is composed of a cellulose-binding molecular module (CBM3a) fused at its N-terminal part to a biotinylated molecule-binding head (monomeric streptavidin) and at its C-terminal part to an unnatural amino acid (UnAA) 4-azido-L-phenylalanine moiety. Combining molecular modeling, synthetic biology and chemistry, we proved that Cerberus is a valuable and versatile platform allowing to functionalize cellulose with organic and inorganic compounds.
We can also plan to associate carbon nanotubes and graphene to AzF head in the aim to change cellulose fibre rigidity and create conductive cellulose, respectively. In order to do that, we successfully activated these compounds by diazotization reaction. The click chemistry assay and validation protocol need improvement to obtain our goal.
Bacterial cellulose has unique properties such as its natural purity may give it a bright future in the industries, that is why we decided to produce functionalized bacterial cellulose in vivo in reactor. As a first step towards the production of functionalized bacterial cellulose in a reactor, we demonstrate that CBM3a module of Cerberus fused to Red fluorescent protein is able to make bacterial cellulose fluorescent. Moreover, our first production of Orthos in Pichia pastoris are very conclusive, but we have to optimize the production.
We truly feel that Cerberus platform is a very convenient and efficient solution to facilitate molecular work for iGEM projects and laboratory work in general. In fact, we can combine the use of two binding sites to purify simply molecules of interest (cellulose/streptavidin or cellulose/AzF or streptavidin/AzF). For example, we can imagine purify cells via biotinylated membrane proteins. An addition of Cerberus protein can permit the interaction between the monomeric streptavidin of Cerberus and the biotin of protein. Then, two possibilities are open to us to purify using either cellulose or paramagnetic beads. After attraction, we can release molecule of interest by using the TEV protease cleavage.
Finally, we can also imagine to change our CBM3a to another proteic domain to associate functionalizing proteins to new materials as plastic to facilitate its degradation for example.
A workflow to produce functionalized cellulose
This project was not only about functionalizing cellulose but ambitioned to define the whole process from component purification to production in vivo (Figure 1).

Indeed, we have defined and validated the basis to model our platform. This allows testing in silico any further modifications that could be of interest for Cerberus. Next, we demonstrated the efficiency of the Cerberus production in E. coli. This validated our choices for the strain, vector and culture conditions (IPTG, AzF, arabinose…). Then, we also successfully purified Cerberus and shown this through SDS-PAGE and Western Blot. As previously described, production of biotinylated compounds was validated as well as cellulose functionalization using Cerberus. We went even further and started the production of cellulose in vivo with Gluconacetobacter hansenii. The next step would have been to demonstrate the possibility to co-culture this strain and a strain synthesizing Cerberus and the function. Then, the final stage will be to start the production of cellulose in vivo. Even if we did not have enough time to each this final step, it was thought from the beginning of the project, as it was for this whole workflow.
No dogs were harmed over the course of this iGEM project.
The whole Toulouse INSA-UPS team wants to thank our sponsors, especially:









And many more. For futher information about our sponsors, please consult our Sponsors page.
The content provided on this website is the fruit of the work of the Toulouse INSA-UPS iGEM Team. As a deliverable for the iGEM Competition, it falls under the Creative Commons Attribution 4.0. Thus, all content on this wiki is available under the Creative Commons Attribution 4.0 license (or any later version). For futher information, please consult the official website of Creative Commons.
This website was designed with Bootstrap (4.1.3). Bootstrap is a front-end library of component for html, css and javascript. It relies on both Popper and jQuery. For further information, please consult the official website of Bootstrap.

