| Line 987: | Line 987: | ||
<ul> | <ul> | ||
<li>Pick 3 colonies and seed then in 9ml LB + Your favourite antibiotic. Incubate O/N at 37°C under 130 rpm shaking.</li> | <li>Pick 3 colonies and seed then in 9ml LB + Your favourite antibiotic. Incubate O/N at 37°C under 130 rpm shaking.</li> | ||
| − | <li>Autoclave 800ml LB 10 g/L NaCl in a 2L baffled flask and | + | <li>Autoclave 800ml LB 10 g/L NaCl in a 2L baffled flask and preheat it at 37°C O/N.</li> |
</ul> | </ul> | ||
</li> | </li> | ||
| Line 994: | Line 994: | ||
<li>Take a sample of the 800ml culture and measure OD600 as blank.</li> | <li>Take a sample of the 800ml culture and measure OD600 as blank.</li> | ||
<li>Take 1ml of preculture and measure OD600, seed the preculture in the 800ml LB and take OD.</li> | <li>Take 1ml of preculture and measure OD600, seed the preculture in the 800ml LB and take OD.</li> | ||
| − | <li>Calculate | + | <li>Calculate OD600 assuming that it doubles every 20 minutes and check the OD until it reaches 0.5 ~0.6. Once the exponontial phase is reached, induce with 800µl of 1M IPTG stock solution and incubate at 37°C for 4h or O/N at 16°C.</li> |
<li>Pellet cells by centrifugate at 5000 x g for 10 minutes and resuspend in 10ml TB 1x 10mM Imidazole if growth was performed at 37°C of 10ml of O/N.</li> | <li>Pellet cells by centrifugate at 5000 x g for 10 minutes and resuspend in 10ml TB 1x 10mM Imidazole if growth was performed at 37°C of 10ml of O/N.</li> | ||
<li>Freeze cells for later purification or wait at least 30 minutes to continue the protocol.</li> | <li>Freeze cells for later purification or wait at least 30 minutes to continue the protocol.</li> | ||
<li>Sonication : Run on cycle of 1 minute and one of 30 seconds, both at 25 of intensity.</li> | <li>Sonication : Run on cycle of 1 minute and one of 30 seconds, both at 25 of intensity.</li> | ||
| − | <li>Pellet cells by centrifugate at | + | <li>Pellet cells by centrifugate at 60,000 x g for 30 minutes and withdraw the supernatant that is the <strong>cell free extract</strong>.</li> |
<li>Then continue to the protein purification protocol.</li> | <li>Then continue to the protein purification protocol.</li> | ||
</ul> | </ul> | ||
| Line 1,017: | Line 1,017: | ||
<hr/> | <hr/> | ||
<h4 id="imac-column-purification" class="heavy">1 . 1 .IMAC column purification</h4> | <h4 id="imac-column-purification" class="heavy">1 . 1 .IMAC column purification</h4> | ||
| − | <p>The volumes will be expressed as CV ( | + | <p>The volumes will be expressed as CV (Column Volume) which refers to the volume of resin deposited.</p> |
<p><strong>Materials</strong></p> | <p><strong>Materials</strong></p> | ||
<ul> | <ul> | ||
Revision as of 21:05, 16 October 2018
EXPERIMENTS
Here, you find all the necessary information about the wetlab materials and methods we used in the course of our project.
Escherichia coli vectors
pET28
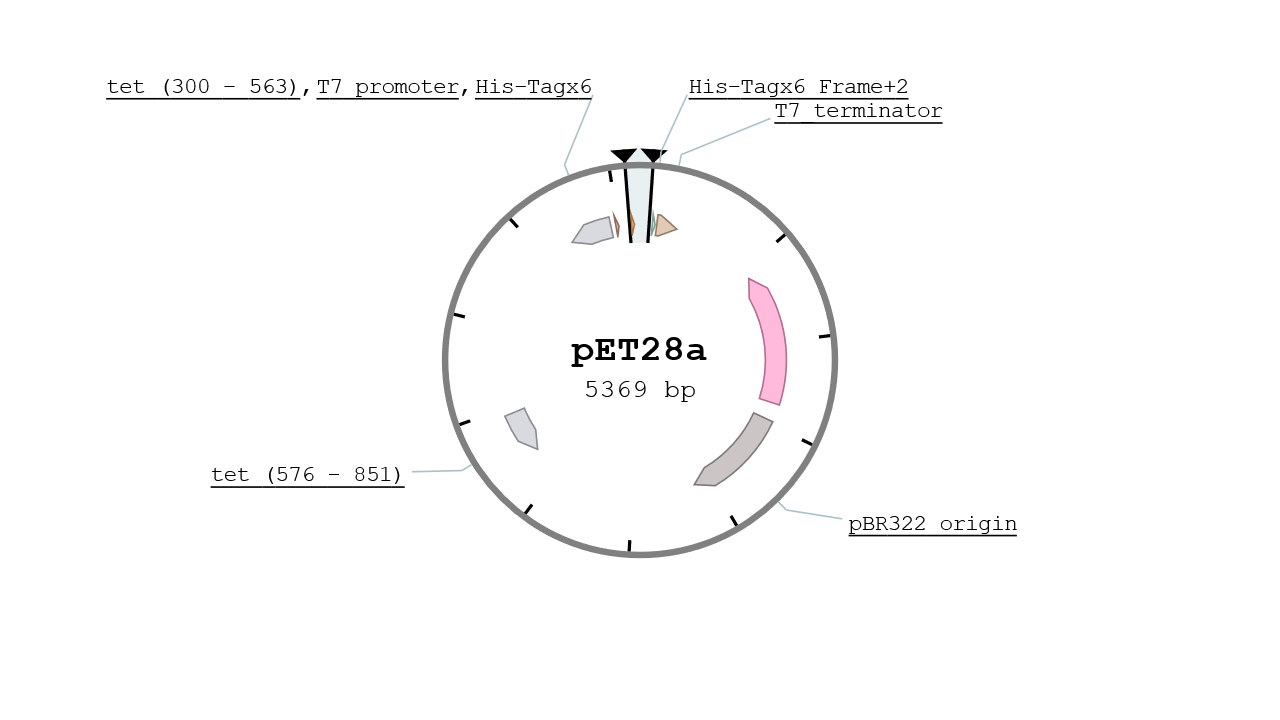
The pET28 vector contains two His Tags in his MCS and is specified for protein expression at high yield. It give the strain a resistance to Kanamycin and is replication origin is the pBR322 which is a mid - low copy number plasmid (~10). The vector used was a kind gift from IBCG library.
pETDuet-1
The pETDuet-1 is, as the pET28, specialized in high yield protein production. It main difference is that it contains two MCS and can therefore express two proteins at the same time under the same expression conditions. It bears ampicilin resistance and is a mid - low copy number plasmid. The plasmid used was a gift from IBCG library.

Pichia pastoris vectors
pPICZ alpha
The pPICZalpha i an integrative plasmid for Pichia pastoris protein expression and suitable for E. coli replication. Its MCS is suitable for protein expession using the methanol inductible AOX1 Pomoter (PAOX1) and offering the alpha factor secretion signal for protein purification from culture supernatant. It contains a His tag and a Myc Tag for affinity purification stategies.

pGAPZalpha
The pGAPZalpha is roughly the same as the pPICZalpha with the difference of the promoter that is GAP Promoter, a glucose indcutible promoter.

Primer used
- Cerberus Forward : TAAGAAGGAGATATACCATGGCGGAAGCGGGTATCACC
- Cerberus Reverse : CTCGAGTGCGGCCGCAAGCTTCGGATCGTCCTATGATGGAGG
- Sirius Forward: TAAGAAGGAGATATACCATGAATGCTACGCCAACTAAGGGTGC
- Sirius Reverse: CTCGAGTGCGGCCGCAAGCTTAGCACCGGTGGAGTGACG
- BirA Forward: AAGGAGATATACATATGAAGGATAACACCGTGCCACTGA
- BirA Reverse: CTTTACCAGACTCGATTATTTTTCTGCACTACGCAGGGA
- BFP Forward: ACCACAGCCAGGATCCTATGAGCGAACTGATCAAAGAGAACA
- BFP Reverse: ATGCGGCCGCAAGCTTCTCATGCCATTCAATTTTCTGTGCT
- RFP Forward: AGGAGATATACCATGGCTTCCTCCGAAGACGTTATCAAAG
- RFP Reverse: GTGCGGCCGCAAGCTTAGCACCGGTGGAGTGACG
- Scygonadin Forward: GGCTGAAGCTGAATTCGGCCAGGCACTCAACAAAC
- Scygonadin Reverse: TGGGCCACGTGAATTCTCACTCATGCCATTCAATTTTCTG
Escherichia coli strains
Plasmid amplification strains- Stellar F-, endA1, supE44, thi-1, recA1, relA1, gyrA96, phoA, Φ80d lacZΔ M15, Δ(lacZYA-argF) U169, Δ(mrr-hsdRMS-mcrBC), ΔmcrA, λ-
- Top10 F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ-
- BL21(DE3) B F-ompT gal dcm lon hsdSB(rB-mB-) λ(DE3[lacI lacUV5-T7p07 ind1 sam7 nin5])[malB+]K-12(λS)
- Tuner F- ompT hsdSB (rB- mB-) gal dcm lacY1(DE3)
Pichia pastoris strains
For protein production we used GS200(ΔARG4, ΔHIS4) Strain for Cerberus production and X33(wt) for Scygonadin
Gluconacetobacter hansenii strain ATCC 53582
Click reagants
- DBCO-Biotin, Sigma CAS : 1255942-07-4
- DBCO-Fluorescein, Jena Bioscience Cat. No. : CLK-051-1
- DBCO-Magnetic beads, Jena Bioscience Cat. No. : CLK-1037-1
- Fluorescein-Azide, Jena Bioscience Cat. No. : CLK-80101-5
- 4-L-azidophenylanaline, Sigma Ref. : 06162
Graphene Functionalization
- Graphene, Sigma Ref. : 900561
- 4-Ethynylaniline, Sigma Ref. : 481122
- Isopentyl Nitrite, Sigma Ref. : 150495
- N,N-Dimathylformamide, Sigma Ref. : 227056
Protein purification
- Cobalt Affinity Gel, Sigma Ref. : H8162
- Protease inhibitor cocktail tablet, Sigma Ref. : S8830
Cellulose Binding assay
- Avicel, Sigma Ref. : 11365
No dogs were harmed over the course of this iGEM project.
The whole Toulouse INSA-UPS team wants to thank our sponsors, especially:









And many more. For futher information about our sponsors, please consult our Sponsors page.
The content provided on this website is the fruit of the work of the Toulouse INSA-UPS iGEM Team. As a deliverable for the iGEM Competition, it falls under the Creative Commons Attribution 4.0. Thus, all content on this wiki is available under the Creative Commons Attribution 4.0 license (or any later version). For futher information, please consult the official website of Creative Commons.
This website was designed with Bootstrap (4.1.3). Bootstrap is a front-end library of component for html, css and javascript. It relies on both Popper and jQuery. For further information, please consult the official website of Bootstrap.

